6.2 Episodic Generalization Optimization - EGO#
Introduction#
Human cognition is unique in its ability to perform a wide range of tasks and to learn new tasks quickly. Both abilities have long been associated with the acquisition of knowledge that can generalize across tasks and the flexible use of that knowledge to execute goal-directed behavior. In this tutorial, we introduce how this can emerge in a neural network by implementing the Episodic Generalization and Optimization (EGO) framework. The framework consists of an episodic memory module, which rapidly learns relationships between stimuli; a semantic pathway, which more slowly learns how stimuli map to responses; and a recurrent context module, which maintains a representation of task-relevant context information, integrates this over time, and uses it to recall context-relevant memories.
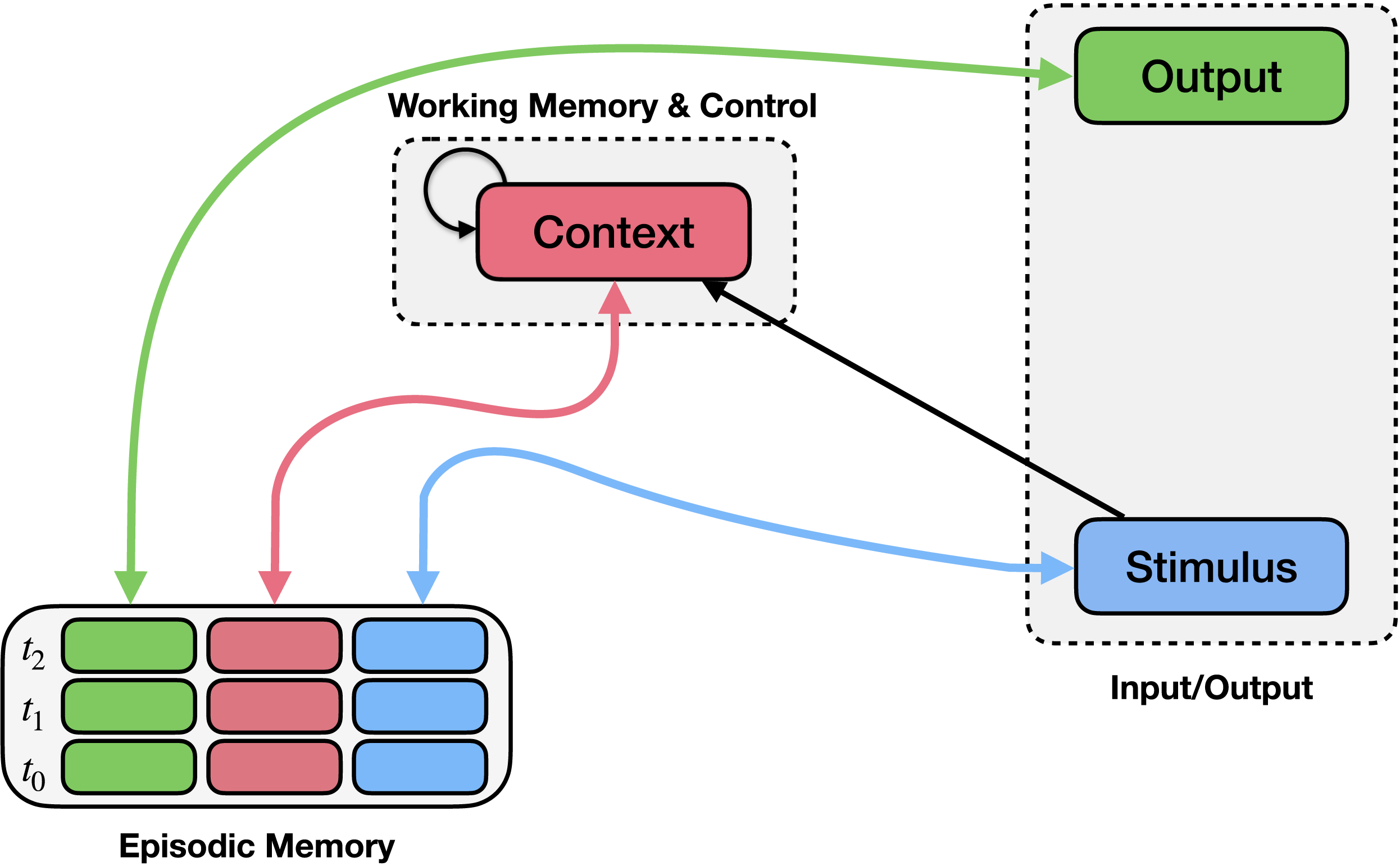
The EGO framework consists of a control mechanism (context module; upper middle) and an episodic memory mechanism (bottom left). Episodic memory records conjunctions of stimuli (blue boxes), contexts (pink boxes), and observed responses (green boxes) at each time point (rows). Bidirectional arrows connect episodic memory to the stimulus, context, and output, indicating that these values can be stored in or used to query episodic memory, or retrieved from it when another field is queried. You can think of this as a more flexible dictionary that stores triplets instead of distinct key-value pairs, and allows any field (or any combinations of fields) to act as a key. The context module integrates previous context (recurrent connection) along with information about the stimulus and the context retrieved from memory.
Here we show that the EGO framework can emulate human behavior in a specific learning environment where participants are trained on two sets of sequences involving identical states presented in different orders for different contexts. Empirical findings show that participants perform better when trained in blocks of each context than when trained interleaved:
Task: Coffe Shop World (CSW)#


Imagine, you are in a city with two coffee shops, each with a different layout and different ways of ordering. In one coffee shop—called The Suspicious Barista—you order first, pay for the coffee, and then sit down to wait until the waiter brings your order. In the other coffee shop—called Café Gratitude—you sit down first, wait until the waiter comes and takes your order. You pay after finishing the coffee.
This example demonstrates that many situations share similar stimuli but have different transition structures. Simple integration will help the system learn the transition structure, but it will only provide a weak cue about the difference between them due to the similarity between the situations. In other words the states –ordering, paying, and sitting down– are very similar between the two situations and are therefore hard to distinguish. This can be overcome by differentiating the context representations associated with each setting (e.g., learning different context representations for coffee shops with paranoid vs. gullible baristas). Recent empirical work suggests that people can learn how to do this very effectively, but that this depends on the temporal structure of the environment: people do better when trained in blocks of each situation than when trained interleaved (Beukers et al., 2023).
We start with creating a dataset for the CSW task.
Installation and Setup
If the following cell fails to execute, please restart the kernel (or session) and run the cell again. This is a known issue when running in google colab.
%%capture
%pip install psyneulink
import psyneulink as pnl
import random
Generating data for the CSW task#
We start by generating a dataset for the CSW task. The dataset consists of sequences of states. The task is to predict the next state given the current state and the context. The transition between states is determined by the context which in turn is determined by the “first” state in the sequence. The following figure illustrates the task structure:
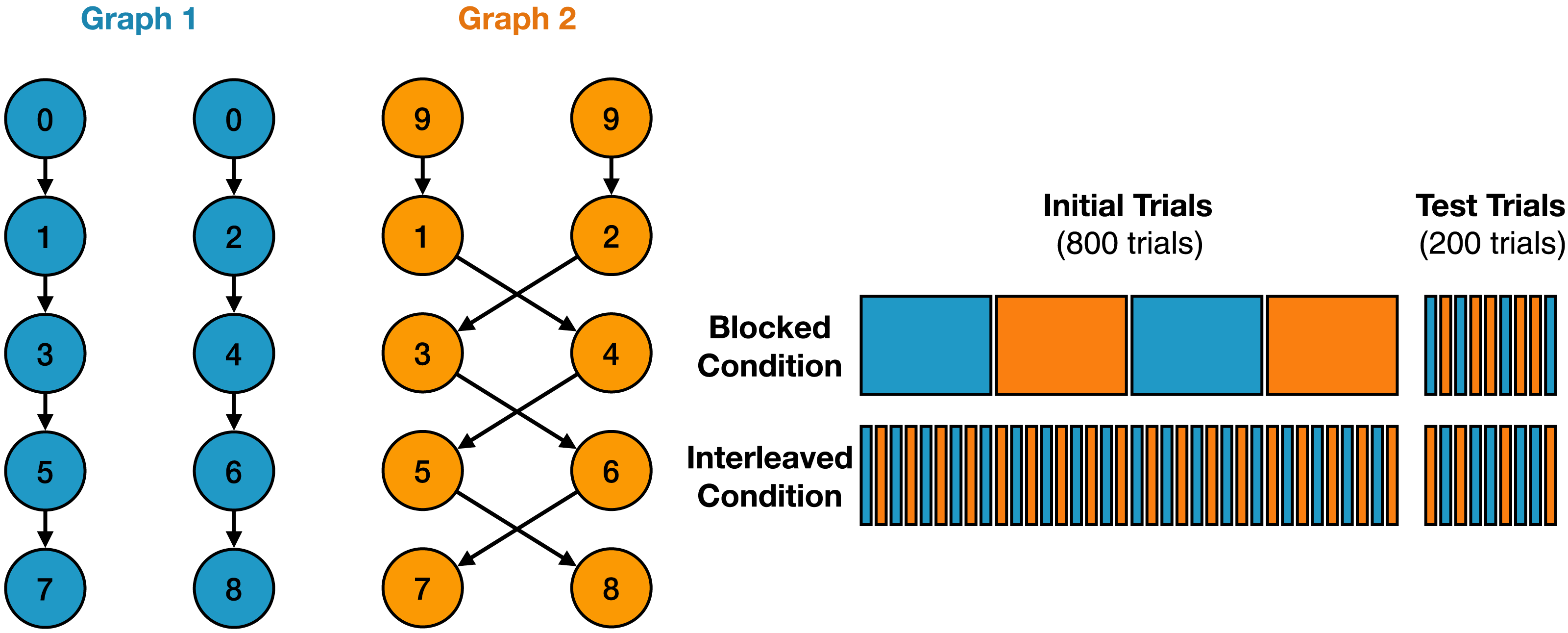
On the left side of the figure, you can see the task structure:
The two colors represent different contexts: blue and orange.
If the first observed state in a sequence is 0, the participant is in the blue context.
The next state can be either 1 or 2.
From then on, transitions are deterministic:
1 → 3 → 5 → 7
2 → 4 → 6 → 8
If the first observed state is 9, the participant is in the orange context.
The sequence starts with either 1 or 2, but follows a different transition pattern:
1 → 4 → 5 → 8
2 → 3 → 6 → 7
The right side of the figure shows the different learning paradigms:
In the blocked paradigm, participants are trained on blocks of the same context. In the interleaved paradigm, participants are trained on a mix of contexts. In the test paradigm, participants are tested on a sequence of random contexts.
We start with defining a function that generates a context-specific sequence:
def gen_context(
context: int,
start_state: int,
):
"""
Generate a context-specific sequence.
Args:
context (int): The context to generate the sequence for. (0 or 9)
start_state (int): The first state in the sequence. (1 or 2)
"""
seq = [context, start_state]
if context == 0:
for _ in range(3):
seq.append(seq[-1] + 2)
elif context == 9:
for _ in range(3):
seq.append(seq[-1] + 1 if seq[-1] % 2 == 0 else seq[-1] + 3)
return seq
"""Test the function"""
assert gen_context(0, 1) == [0, 1, 3, 5, 7]
assert gen_context(9, 2) == [9, 2, 3, 6, 7]
Generate a full dataset for the CSW task. Now, let’s create a function that returns the full trial sequence for a given paradigm and number of samples.
# Define the paradigms
BLOCKED = 'blocked'
INTERLEAVED = 'interleaved'
def gen_context_sequences(
paradigm: str,
train_contexts: int,
test_contexts: int,
block_size: int = 4,
):
"""
Generate a dataset for the CSW task.
Args:
paradigm (str): The paradigm to generate the dataset for. (blocked or interleaved)
train_contexts (int): The number of training contexts.
test_contexts (int): The number of test contexts.
block_size (int): The size of each block in the blocked paradigm.
"""
assert train_contexts % block_size == 0, "The number of training samples must be a multiple of block_size."
x = []
if paradigm == INTERLEAVED:
for idx in range(train_contexts):
if idx % 2: # odd contexts -> context 0
x += [gen_context(0, random.randint(1, 2))]
else: # even contexts -> context 9
x += [gen_context(9, random.randint(1, 2))]
if paradigm == BLOCKED:
for i in range(block_size): # block_size number of blocks
if i % 2: # odd blocks -> context 0
for _ in range(train_contexts // block_size):
x += [gen_context(0, random.randint(1, 2))]
else: # even blocks -> context 9
for _ in range(train_contexts // block_size):
x += [gen_context(9, random.randint(1, 2))]
for _ in range(test_contexts):
x += [gen_context(random.choice([0, 9]), random.randint(1, 2))]
return x
context_sequences = gen_context_sequences(BLOCKED, 8, 4)
context_sequences
[[9, 2, 3, 6, 7],
[9, 1, 4, 5, 8],
[0, 1, 3, 5, 7],
[0, 2, 4, 6, 8],
[9, 2, 3, 6, 7],
[9, 1, 4, 5, 8],
[0, 1, 3, 5, 7],
[0, 2, 4, 6, 8],
[9, 1, 4, 5, 8],
[9, 2, 3, 6, 7],
[9, 1, 4, 5, 8],
[9, 2, 3, 6, 7]]
The structure of the generated sequence is not “realistic” yet. The participant doesn’t see distinct contexts but rather states. We need to “flatten” the sequence. Also, we instead of using integers to represent the states, we will use one-hot encoding:
def one_hot_encode(
label: int,
num_classes: int):
"""
One hot encode a label (integer)
Args:
label (int): The label to encode (between 0 and num_classes-1)
num_classes (int): The number of classes
"""
return [1 if i == label else 0 for i in range(num_classes)]
def state_sequence(
paradigm: str,
train_trials: int,
test_trials: int,
context_length: int = 5,
block_size: int = 4,
):
"""
Generate a dataset for the CSW task.
Args:
paradigm (str): The paradigm to generate the dataset for. (blocked or interleaved)
train_trials (int): The number of training trials.
test_trials (int): The number of test trials.
context_length (int): The length of the context.
block_size (int): The size of each block in the blocked paradigm.
"""
assert train_trials % context_length == 0, "The number of training samples must be a multiple of context_length."
assert test_trials % context_length == 0, "The number of test samples must be a multiple of context_length."
train_contexts = train_trials // context_length
test_contexts = test_trials // context_length
train_context_sequences = gen_context_sequences(
paradigm, train_contexts, test_contexts, block_size
)
states = []
for context_sequence in train_context_sequences:
for state_int in context_sequence:
states.append(one_hot_encode(state_int, 11))
return states
state_sequences = state_sequence(BLOCKED, 20, 5)
state_sequences
[[0, 0, 0, 0, 0, 0, 0, 0, 0, 1, 0],
[0, 0, 1, 0, 0, 0, 0, 0, 0, 0, 0],
[0, 0, 0, 1, 0, 0, 0, 0, 0, 0, 0],
[0, 0, 0, 0, 0, 0, 1, 0, 0, 0, 0],
[0, 0, 0, 0, 0, 0, 0, 1, 0, 0, 0],
[1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0],
[0, 0, 1, 0, 0, 0, 0, 0, 0, 0, 0],
[0, 0, 0, 0, 1, 0, 0, 0, 0, 0, 0],
[0, 0, 0, 0, 0, 0, 1, 0, 0, 0, 0],
[0, 0, 0, 0, 0, 0, 0, 0, 1, 0, 0],
[0, 0, 0, 0, 0, 0, 0, 0, 0, 1, 0],
[0, 1, 0, 0, 0, 0, 0, 0, 0, 0, 0],
[0, 0, 0, 0, 1, 0, 0, 0, 0, 0, 0],
[0, 0, 0, 0, 0, 1, 0, 0, 0, 0, 0],
[0, 0, 0, 0, 0, 0, 0, 0, 1, 0, 0],
[1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0],
[0, 0, 1, 0, 0, 0, 0, 0, 0, 0, 0],
[0, 0, 0, 0, 1, 0, 0, 0, 0, 0, 0],
[0, 0, 0, 0, 0, 0, 1, 0, 0, 0, 0],
[0, 0, 0, 0, 0, 0, 0, 0, 1, 0, 0],
[0, 0, 0, 0, 0, 0, 0, 0, 0, 1, 0],
[0, 1, 0, 0, 0, 0, 0, 0, 0, 0, 0],
[0, 0, 0, 0, 1, 0, 0, 0, 0, 0, 0],
[0, 0, 0, 0, 0, 1, 0, 0, 0, 0, 0],
[0, 0, 0, 0, 0, 0, 0, 0, 1, 0, 0]]
Why do we encode the states using one-hot encoding?
✅ Solution 1
One-hot encoding is used for categorical variables. This means states have no inherit “order” or can be compared using arithmetic operations in a meaningful way. One-hot encoding allows this representation as states are “orthogonal” to each other.
We want to train the EGO model in a supervised manner but the generated dataset doesn’t allow us to do so. Why is this the case and what do we need to do be able to train the model?
💡 Hint 1
For supervised training, we need to provide a target for each input. Think about what the target should be in this case.
💡 Hint 2
The task in this case, is to predict the next state given the current state.
✅ Solution 2
The target in this case is just the next state in the sequence:
x = state_sequence(BLOCKED, 20, 5)
y = x[1:] + [one_hot_encode(0, 11)] # the last state has no next state and is arbitrary in this case either 0 or 9
The EGO model#
As mentioned earlier, the EGO model consists of three main components: an episodic memory module, a semantic pathway, and a recurrent context module. PsyNeulink provides a EMComposition class that allows us to create the episodic memory module. The EMcomposition class is a subclass of the Composition class. A strength of the PsyNeuLink framework is that it allows fo the creation of complex composition that can be used as mechanism in other compositions. Here, we first look at the EMComposition class in isolation and then integrate it into the EGO model.
Episodic Memory Module - EMComposition#
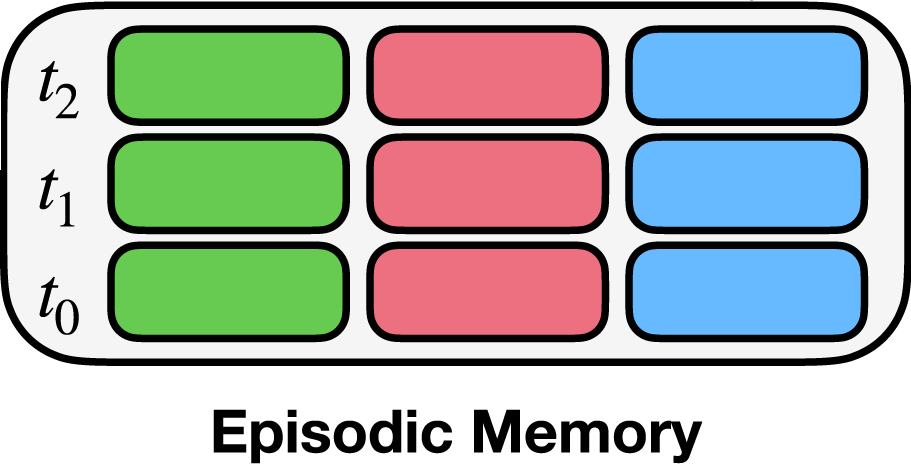
Here, we initialize the EMComposition for the episodic memory shown above. The EMComposition allows for specifying the structure of the episodic memory. Remember, the task here is to predict the state from the previous state and the context. Therefore, in our case each entry in the memory consists of a triplet of states:
The current state (green box)
The previous state (blue box)
The context (pink box)
Each state is represented as a vector with 11 elements (one hot encoding).
Here, we also specify the specific fields. Fields have three main parameters that have to be specified as a dictionary:
FIELD_WEIGHT: The weight of the field when retrieving from memoryLEARN_FIELD_WEIGHT: Whether the retrieval field weight should be learned (Here, we won’t learn these weights but set them)TARGET_FIELD: Whether the field is a target field (Meaning it’s “error” is calculated during learning)
Before looking at the code below, think about what to set for the FIELD_WEIGHT and the TARGET_FIELD for the three different fields (current state, previous state, and context).
💡 Hint
The FIELD_WEIGHT specifies weather a field should be used during retrieval (and how much it should be used during retrieval). It is a scalar value between 0 and 1. The TARGET_FIELD specifies weather a field is a target field.
✅ Solution
The FIELD_WEIGHT for the current state should be None since it is the target field and shouldn’t be used in retrieval. The FIELD_WEIGHT for both the previous and the context should be set to an equal value (here we set them both to 1). The TARGET_FIELD should be set to True for the current state and False for the previous state and the context.
name = 'EM' # a name for the EMComposition
# Memory parameters
state_size = 11 # the size of the state vector
memory_capacity = 1000 # here we set the maximum number of entries in the memory (we want to be able to store all 1000 trials)
# Fields
# State field
state_name = 'STATE'
state_retrieval_weight = None # This entry is not used when retrieving from memory (remember, we want to predict the state)
state_is_target = True
# Previous state field
previous_state_name = 'PREVIOUS STATE'
previous_state_retrieval_weight = .5 # This entry is used when retrieving from memory
previous_state_is_target = False
# Context field
context_name = 'CONTEXT'
context_retrieval_weight = .5 # This entry is used when retrieving from memory
context_is_target = False
em = pnl.EMComposition(name=name,
memory_template=[[0] * state_size, # state
[0] * state_size, # previous state
[0] * state_size], # context
memory_fill=.001,
memory_capacity=memory_capacity,
normalize_memories=False,
memory_decay_rate=0, # no decay of memory
softmax_gain=10.,
softmax_threshold=.001,
fields={state_name: {pnl.FIELD_WEIGHT: state_retrieval_weight,
pnl.LEARN_FIELD_WEIGHT: False,
pnl.TARGET_FIELD: True},
previous_state_name: {pnl.FIELD_WEIGHT: previous_state_retrieval_weight,
pnl.LEARN_FIELD_WEIGHT: False,
pnl.TARGET_FIELD: False},
context_name: {pnl.FIELD_WEIGHT: context_retrieval_weight,
pnl.LEARN_FIELD_WEIGHT: False,
pnl.TARGET_FIELD: False}},
normalize_field_weights=True,
concatenate_queries=False,
enable_learning=True,
learning_rate=.5,
device=pnl.CPU
)
Let’s see how the EMComposition looks like:
em.show_graph(output_fmt='jupyter')
---------------------------------------------------------------------------
FileNotFoundError Traceback (most recent call last)
File /opt/hostedtoolcache/Python/3.11.14/x64/lib/python3.11/site-packages/graphviz/backend/execute.py:76, in run_check(cmd, input_lines, encoding, quiet, **kwargs)
75 kwargs['stdout'] = kwargs['stderr'] = subprocess.PIPE
---> 76 proc = _run_input_lines(cmd, input_lines, kwargs=kwargs)
77 else:
File /opt/hostedtoolcache/Python/3.11.14/x64/lib/python3.11/site-packages/graphviz/backend/execute.py:96, in _run_input_lines(cmd, input_lines, kwargs)
95 def _run_input_lines(cmd, input_lines, *, kwargs):
---> 96 popen = subprocess.Popen(cmd, stdin=subprocess.PIPE, **kwargs)
98 stdin_write = popen.stdin.write
File /opt/hostedtoolcache/Python/3.11.14/x64/lib/python3.11/subprocess.py:1026, in Popen.__init__(self, args, bufsize, executable, stdin, stdout, stderr, preexec_fn, close_fds, shell, cwd, env, universal_newlines, startupinfo, creationflags, restore_signals, start_new_session, pass_fds, user, group, extra_groups, encoding, errors, text, umask, pipesize, process_group)
1023 self.stderr = io.TextIOWrapper(self.stderr,
1024 encoding=encoding, errors=errors)
-> 1026 self._execute_child(args, executable, preexec_fn, close_fds,
1027 pass_fds, cwd, env,
1028 startupinfo, creationflags, shell,
1029 p2cread, p2cwrite,
1030 c2pread, c2pwrite,
1031 errread, errwrite,
1032 restore_signals,
1033 gid, gids, uid, umask,
1034 start_new_session, process_group)
1035 except:
1036 # Cleanup if the child failed starting.
File /opt/hostedtoolcache/Python/3.11.14/x64/lib/python3.11/subprocess.py:1955, in Popen._execute_child(self, args, executable, preexec_fn, close_fds, pass_fds, cwd, env, startupinfo, creationflags, shell, p2cread, p2cwrite, c2pread, c2pwrite, errread, errwrite, restore_signals, gid, gids, uid, umask, start_new_session, process_group)
1954 if err_filename is not None:
-> 1955 raise child_exception_type(errno_num, err_msg, err_filename)
1956 else:
FileNotFoundError: [Errno 2] No such file or directory: PosixPath('dot')
The above exception was the direct cause of the following exception:
ExecutableNotFound Traceback (most recent call last)
File /opt/hostedtoolcache/Python/3.11.14/x64/lib/python3.11/site-packages/IPython/core/formatters.py:1036, in MimeBundleFormatter.__call__(self, obj, include, exclude)
1033 method = get_real_method(obj, self.print_method)
1035 if method is not None:
-> 1036 return method(include=include, exclude=exclude)
1037 return None
1038 else:
File /opt/hostedtoolcache/Python/3.11.14/x64/lib/python3.11/site-packages/graphviz/jupyter_integration.py:98, in JupyterIntegration._repr_mimebundle_(self, include, exclude, **_)
96 include = set(include) if include is not None else {self._jupyter_mimetype}
97 include -= set(exclude or [])
---> 98 return {mimetype: getattr(self, method_name)()
99 for mimetype, method_name in MIME_TYPES.items()
100 if mimetype in include}
File /opt/hostedtoolcache/Python/3.11.14/x64/lib/python3.11/site-packages/graphviz/jupyter_integration.py:98, in <dictcomp>(.0)
96 include = set(include) if include is not None else {self._jupyter_mimetype}
97 include -= set(exclude or [])
---> 98 return {mimetype: getattr(self, method_name)()
99 for mimetype, method_name in MIME_TYPES.items()
100 if mimetype in include}
File /opt/hostedtoolcache/Python/3.11.14/x64/lib/python3.11/site-packages/graphviz/jupyter_integration.py:112, in JupyterIntegration._repr_image_svg_xml(self)
110 def _repr_image_svg_xml(self) -> str:
111 """Return the rendered graph as SVG string."""
--> 112 return self.pipe(format='svg', encoding=SVG_ENCODING)
File /opt/hostedtoolcache/Python/3.11.14/x64/lib/python3.11/site-packages/graphviz/piping.py:104, in Pipe.pipe(self, format, renderer, formatter, neato_no_op, quiet, engine, encoding)
55 def pipe(self,
56 format: typing.Optional[str] = None,
57 renderer: typing.Optional[str] = None,
(...) 61 engine: typing.Optional[str] = None,
62 encoding: typing.Optional[str] = None) -> typing.Union[bytes, str]:
63 """Return the source piped through the Graphviz layout command.
64
65 Args:
(...) 102 '<?xml version='
103 """
--> 104 return self._pipe_legacy(format,
105 renderer=renderer,
106 formatter=formatter,
107 neato_no_op=neato_no_op,
108 quiet=quiet,
109 engine=engine,
110 encoding=encoding)
File /opt/hostedtoolcache/Python/3.11.14/x64/lib/python3.11/site-packages/graphviz/_tools.py:185, in deprecate_positional_args.<locals>.decorator.<locals>.wrapper(*args, **kwargs)
177 wanted = ', '.join(f'{name}={value!r}'
178 for name, value in deprecated.items())
179 warnings.warn(f'The signature of {func_name} will be reduced'
180 f' to {supported_number} positional arg{s_}{qualification}'
181 f' {list(supported)}: pass {wanted} as keyword arg{s_}',
182 stacklevel=stacklevel,
183 category=category)
--> 185 return func(*args, **kwargs)
File /opt/hostedtoolcache/Python/3.11.14/x64/lib/python3.11/site-packages/graphviz/piping.py:121, in Pipe._pipe_legacy(self, format, renderer, formatter, neato_no_op, quiet, engine, encoding)
112 @_tools.deprecate_positional_args(supported_number=1, ignore_arg='self')
113 def _pipe_legacy(self,
114 format: typing.Optional[str] = None,
(...) 119 engine: typing.Optional[str] = None,
120 encoding: typing.Optional[str] = None) -> typing.Union[bytes, str]:
--> 121 return self._pipe_future(format,
122 renderer=renderer,
123 formatter=formatter,
124 neato_no_op=neato_no_op,
125 quiet=quiet,
126 engine=engine,
127 encoding=encoding)
File /opt/hostedtoolcache/Python/3.11.14/x64/lib/python3.11/site-packages/graphviz/piping.py:149, in Pipe._pipe_future(self, format, renderer, formatter, neato_no_op, quiet, engine, encoding)
146 if encoding is not None:
147 if codecs.lookup(encoding) is codecs.lookup(self.encoding):
148 # common case: both stdin and stdout need the same encoding
--> 149 return self._pipe_lines_string(*args, encoding=encoding, **kwargs)
150 try:
151 raw = self._pipe_lines(*args, input_encoding=self.encoding, **kwargs)
File /opt/hostedtoolcache/Python/3.11.14/x64/lib/python3.11/site-packages/graphviz/backend/piping.py:212, in pipe_lines_string(engine, format, input_lines, encoding, renderer, formatter, neato_no_op, quiet)
206 cmd = dot_command.command(engine, format,
207 renderer=renderer,
208 formatter=formatter,
209 neato_no_op=neato_no_op)
210 kwargs = {'input_lines': input_lines, 'encoding': encoding}
--> 212 proc = execute.run_check(cmd, capture_output=True, quiet=quiet, **kwargs)
213 return proc.stdout
File /opt/hostedtoolcache/Python/3.11.14/x64/lib/python3.11/site-packages/graphviz/backend/execute.py:81, in run_check(cmd, input_lines, encoding, quiet, **kwargs)
79 except OSError as e:
80 if e.errno == errno.ENOENT:
---> 81 raise ExecutableNotFound(cmd) from e
82 raise
84 if not quiet and proc.stderr:
ExecutableNotFound: failed to execute PosixPath('dot'), make sure the Graphviz executables are on your systems' PATH
<graphviz.graphs.Digraph at 0x7f9abc9b9ed0>
Input, Context, and Output Layers#
Next, we “hook” up the EMComposition to the input, output and context layer.

We start with defining the layers
Before defining the layers, make sure you understand the in and output of the model:
Although the episodic memory composition has three “memory slot”, our training set only consists of a stream of a single state. How can we use this single state
state_input_layer = pnl.ProcessingMechanism(name=state_name, input_shapes=state_size)
previous_state_layer = pnl.ProcessingMechanism(name=previous_state_name, input_shapes=state_size)
context_layer = pnl.TransferMechanism(name=context_name,
input_shapes=state_size,
function=pnl.Tanh,
integrator_mode=True,
integration_rate=.69)
# The output layer:
prediction_layer = pnl.ProcessingMechanism(name='PREDICTION', input_shapes=state_size)
After defining the layers, we need to specify the pathways between the layers. Before looking at the code below, think about which pathways (if any) are learned and which ones are fixed.
# Names for the input nodes of the EMComposition have the form: <node_name> + ' [QUERY]' or <node_name> + ' [VALUE]' or <node_name> + ' [RETRIEVED]' (see above)
QUERY = ' [QUERY]'
VALUE = ' [VALUE]'
RETRIEVED = ' [RETRIEVED]'
# Pathways
state_to_previous_state_pathway = [state_input_layer,
pnl.MappingProjection(matrix=pnl.IDENTITY_MATRIX,
learnable=False),
previous_state_layer]
state_to_context_pathway = [state_input_layer,
pnl.MappingProjection(matrix=pnl.IDENTITY_MATRIX,
learnable=False),
context_layer]
state_to_em_pathway = [state_input_layer,
pnl.MappingProjection(sender=state_input_layer,
receiver=em.nodes[state_name + VALUE],
matrix=pnl.IDENTITY_MATRIX,
learnable=False),
em]
previous_state_to_em_pathway = [previous_state_layer,
pnl.MappingProjection(sender=previous_state_layer,
receiver=em.nodes[previous_state_name + QUERY],
matrix=pnl.IDENTITY_MATRIX,
learnable=False),
em]
context_learning_pathway = [context_layer,
pnl.MappingProjection(sender=context_layer,
matrix=pnl.IDENTITY_MATRIX,
receiver=em.nodes[context_name + QUERY],
learnable=True),
em,
pnl.MappingProjection(sender=em.nodes[state_name + RETRIEVED],
receiver=prediction_layer,
matrix=pnl.IDENTITY_MATRIX,
learnable=False),
prediction_layer]
Now, we can create the composition
learning_rate = .5
loss_spec = pnl.Loss.BINARY_CROSS_ENTROPY
model_name = 'EGO'
device = pnl.CPU
ego_model = pnl.AutodiffComposition([state_to_previous_state_pathway,
state_to_context_pathway,
state_to_em_pathway,
previous_state_to_em_pathway,
context_learning_pathway],
learning_rate=.5,
loss_spec=pnl.Loss.BINARY_CROSS_ENTROPY,
name='EGO',
device=pnl.CPU)
ego_model.show_graph(output_fmt='jupyter')
---------------------------------------------------------------------------
FileNotFoundError Traceback (most recent call last)
File /opt/hostedtoolcache/Python/3.11.14/x64/lib/python3.11/site-packages/graphviz/backend/execute.py:76, in run_check(cmd, input_lines, encoding, quiet, **kwargs)
75 kwargs['stdout'] = kwargs['stderr'] = subprocess.PIPE
---> 76 proc = _run_input_lines(cmd, input_lines, kwargs=kwargs)
77 else:
File /opt/hostedtoolcache/Python/3.11.14/x64/lib/python3.11/site-packages/graphviz/backend/execute.py:96, in _run_input_lines(cmd, input_lines, kwargs)
95 def _run_input_lines(cmd, input_lines, *, kwargs):
---> 96 popen = subprocess.Popen(cmd, stdin=subprocess.PIPE, **kwargs)
98 stdin_write = popen.stdin.write
File /opt/hostedtoolcache/Python/3.11.14/x64/lib/python3.11/subprocess.py:1026, in Popen.__init__(self, args, bufsize, executable, stdin, stdout, stderr, preexec_fn, close_fds, shell, cwd, env, universal_newlines, startupinfo, creationflags, restore_signals, start_new_session, pass_fds, user, group, extra_groups, encoding, errors, text, umask, pipesize, process_group)
1023 self.stderr = io.TextIOWrapper(self.stderr,
1024 encoding=encoding, errors=errors)
-> 1026 self._execute_child(args, executable, preexec_fn, close_fds,
1027 pass_fds, cwd, env,
1028 startupinfo, creationflags, shell,
1029 p2cread, p2cwrite,
1030 c2pread, c2pwrite,
1031 errread, errwrite,
1032 restore_signals,
1033 gid, gids, uid, umask,
1034 start_new_session, process_group)
1035 except:
1036 # Cleanup if the child failed starting.
File /opt/hostedtoolcache/Python/3.11.14/x64/lib/python3.11/subprocess.py:1955, in Popen._execute_child(self, args, executable, preexec_fn, close_fds, pass_fds, cwd, env, startupinfo, creationflags, shell, p2cread, p2cwrite, c2pread, c2pwrite, errread, errwrite, restore_signals, gid, gids, uid, umask, start_new_session, process_group)
1954 if err_filename is not None:
-> 1955 raise child_exception_type(errno_num, err_msg, err_filename)
1956 else:
FileNotFoundError: [Errno 2] No such file or directory: PosixPath('dot')
The above exception was the direct cause of the following exception:
ExecutableNotFound Traceback (most recent call last)
File /opt/hostedtoolcache/Python/3.11.14/x64/lib/python3.11/site-packages/IPython/core/formatters.py:1036, in MimeBundleFormatter.__call__(self, obj, include, exclude)
1033 method = get_real_method(obj, self.print_method)
1035 if method is not None:
-> 1036 return method(include=include, exclude=exclude)
1037 return None
1038 else:
File /opt/hostedtoolcache/Python/3.11.14/x64/lib/python3.11/site-packages/graphviz/jupyter_integration.py:98, in JupyterIntegration._repr_mimebundle_(self, include, exclude, **_)
96 include = set(include) if include is not None else {self._jupyter_mimetype}
97 include -= set(exclude or [])
---> 98 return {mimetype: getattr(self, method_name)()
99 for mimetype, method_name in MIME_TYPES.items()
100 if mimetype in include}
File /opt/hostedtoolcache/Python/3.11.14/x64/lib/python3.11/site-packages/graphviz/jupyter_integration.py:98, in <dictcomp>(.0)
96 include = set(include) if include is not None else {self._jupyter_mimetype}
97 include -= set(exclude or [])
---> 98 return {mimetype: getattr(self, method_name)()
99 for mimetype, method_name in MIME_TYPES.items()
100 if mimetype in include}
File /opt/hostedtoolcache/Python/3.11.14/x64/lib/python3.11/site-packages/graphviz/jupyter_integration.py:112, in JupyterIntegration._repr_image_svg_xml(self)
110 def _repr_image_svg_xml(self) -> str:
111 """Return the rendered graph as SVG string."""
--> 112 return self.pipe(format='svg', encoding=SVG_ENCODING)
File /opt/hostedtoolcache/Python/3.11.14/x64/lib/python3.11/site-packages/graphviz/piping.py:104, in Pipe.pipe(self, format, renderer, formatter, neato_no_op, quiet, engine, encoding)
55 def pipe(self,
56 format: typing.Optional[str] = None,
57 renderer: typing.Optional[str] = None,
(...) 61 engine: typing.Optional[str] = None,
62 encoding: typing.Optional[str] = None) -> typing.Union[bytes, str]:
63 """Return the source piped through the Graphviz layout command.
64
65 Args:
(...) 102 '<?xml version='
103 """
--> 104 return self._pipe_legacy(format,
105 renderer=renderer,
106 formatter=formatter,
107 neato_no_op=neato_no_op,
108 quiet=quiet,
109 engine=engine,
110 encoding=encoding)
File /opt/hostedtoolcache/Python/3.11.14/x64/lib/python3.11/site-packages/graphviz/_tools.py:185, in deprecate_positional_args.<locals>.decorator.<locals>.wrapper(*args, **kwargs)
177 wanted = ', '.join(f'{name}={value!r}'
178 for name, value in deprecated.items())
179 warnings.warn(f'The signature of {func_name} will be reduced'
180 f' to {supported_number} positional arg{s_}{qualification}'
181 f' {list(supported)}: pass {wanted} as keyword arg{s_}',
182 stacklevel=stacklevel,
183 category=category)
--> 185 return func(*args, **kwargs)
File /opt/hostedtoolcache/Python/3.11.14/x64/lib/python3.11/site-packages/graphviz/piping.py:121, in Pipe._pipe_legacy(self, format, renderer, formatter, neato_no_op, quiet, engine, encoding)
112 @_tools.deprecate_positional_args(supported_number=1, ignore_arg='self')
113 def _pipe_legacy(self,
114 format: typing.Optional[str] = None,
(...) 119 engine: typing.Optional[str] = None,
120 encoding: typing.Optional[str] = None) -> typing.Union[bytes, str]:
--> 121 return self._pipe_future(format,
122 renderer=renderer,
123 formatter=formatter,
124 neato_no_op=neato_no_op,
125 quiet=quiet,
126 engine=engine,
127 encoding=encoding)
File /opt/hostedtoolcache/Python/3.11.14/x64/lib/python3.11/site-packages/graphviz/piping.py:149, in Pipe._pipe_future(self, format, renderer, formatter, neato_no_op, quiet, engine, encoding)
146 if encoding is not None:
147 if codecs.lookup(encoding) is codecs.lookup(self.encoding):
148 # common case: both stdin and stdout need the same encoding
--> 149 return self._pipe_lines_string(*args, encoding=encoding, **kwargs)
150 try:
151 raw = self._pipe_lines(*args, input_encoding=self.encoding, **kwargs)
File /opt/hostedtoolcache/Python/3.11.14/x64/lib/python3.11/site-packages/graphviz/backend/piping.py:212, in pipe_lines_string(engine, format, input_lines, encoding, renderer, formatter, neato_no_op, quiet)
206 cmd = dot_command.command(engine, format,
207 renderer=renderer,
208 formatter=formatter,
209 neato_no_op=neato_no_op)
210 kwargs = {'input_lines': input_lines, 'encoding': encoding}
--> 212 proc = execute.run_check(cmd, capture_output=True, quiet=quiet, **kwargs)
213 return proc.stdout
File /opt/hostedtoolcache/Python/3.11.14/x64/lib/python3.11/site-packages/graphviz/backend/execute.py:81, in run_check(cmd, input_lines, encoding, quiet, **kwargs)
79 except OSError as e:
80 if e.errno == errno.ENOENT:
---> 81 raise ExecutableNotFound(cmd) from e
82 raise
84 if not quiet and proc.stderr:
ExecutableNotFound: failed to execute PosixPath('dot'), make sure the Graphviz executables are on your systems' PATH
<graphviz.graphs.Digraph at 0x7f9985561310>
We also need to specify the learning pathway, which can be inferred from the paramaters we have set (setting the target in EMComposition and setting the context to em pathway as learnable):
learning_components = ego_model.infer_backpropagation_learning_pathways(pnl.ExecutionMode.PyTorch)
ego_model.add_projection(pnl.MappingProjection(sender=state_input_layer,
receiver=learning_components[0],
learnable=False))
(MappingProjection MappingProjection from STATE[OutputPort-0] to TARGET for PREDICTION[InputPort-0])
We also have to make sure the em is executed before the previous state and the context layer:
ego_model.scheduler.add_condition(em, pnl.BeforeNodes(previous_state_layer, context_layer))
Now, we are set to run the model:
trials = state_sequence(BLOCKED, 800, 200)
ego_model.learn(inputs={state_name: trials},
learning_rate=.5,
execution_mode= pnl.ExecutionMode.PyTorch,
)
/opt/hostedtoolcache/Python/3.11.14/x64/lib/python3.11/site-packages/psyneulink/core/components/functions/nonstateful/transferfunctions.py:3499: UserWarning: Softmax function: mask_threshold is set to tensor([0.0010], dtype=torch.float64), but input contains negative values. Masking will be applied to the magnitude of the input.
warnings.warn(f"Softmax function: mask_threshold is set to {mask_threshold}, "
---------------------------------------------------------------------------
KeyboardInterrupt Traceback (most recent call last)
Cell In[12], line 3
1 trials = state_sequence(BLOCKED, 800, 200)
----> 3 ego_model.learn(inputs={state_name: trials},
4 learning_rate=.5,
5 execution_mode= pnl.ExecutionMode.PyTorch,
6 )
File /opt/hostedtoolcache/Python/3.11.14/x64/lib/python3.11/site-packages/psyneulink/core/globals/context.py:747, in handle_external_context.<locals>.decorator.<locals>.wrapper(context, *args, **kwargs)
744 pass
746 try:
--> 747 return func(*args, context=context, **kwargs)
748 except TypeError as e:
749 # context parameter may be passed as a positional arg
750 if (
751 f"{func.__name__}() got multiple values for argument"
752 not in str(e)
753 ):
File /opt/hostedtoolcache/Python/3.11.14/x64/lib/python3.11/site-packages/psyneulink/library/compositions/autodiffcomposition.py:1761, in AutodiffComposition.learn(self, synch_projection_matrices_with_torch, synch_node_variables_with_torch, synch_node_values_with_torch, synch_results_with_torch, retain_torch_trained_outputs, retain_torch_targets, retain_torch_losses, context, base_context, skip_initialization, *args, **kwargs)
1756 if execution_mode == pnlvm.ExecutionMode.PyTorch and not torch_available:
1757 raise AutodiffCompositionError(f"'{self.name}.learn()' has been called with ExecutionMode.Pytorch, "
1758 f"but Pytorch module ('torch') is not installed. "
1759 f"Please install it with `pip install torch` or `pip3 install torch`")
-> 1761 return super().learn(*args,
1762 synch_with_pnl_options=synch_with_pnl_options,
1763 retain_in_pnl_options=retain_in_pnl_options,
1764 execution_mode=execution_mode,
1765 context=context,
1766 base_context=base_context,
1767 skip_initialization=skip_initialization,
1768 **kwargs)
File /opt/hostedtoolcache/Python/3.11.14/x64/lib/python3.11/site-packages/psyneulink/core/globals/context.py:747, in handle_external_context.<locals>.decorator.<locals>.wrapper(context, *args, **kwargs)
744 pass
746 try:
--> 747 return func(*args, context=context, **kwargs)
748 except TypeError as e:
749 # context parameter may be passed as a positional arg
750 if (
751 f"{func.__name__}() got multiple values for argument"
752 not in str(e)
753 ):
File /opt/hostedtoolcache/Python/3.11.14/x64/lib/python3.11/site-packages/psyneulink/core/compositions/composition.py:12101, in Composition.learn(self, inputs, targets, num_trials, epochs, learning_rate, minibatch_size, optimizations_per_minibatch, patience, min_delta, execution_mode, randomize_minibatches, call_before_minibatch, call_after_minibatch, context, base_context, skip_initialization, *args, **kwargs)
12098 if optimizations_per_minibatch is None:
12099 optimizations_per_minibatch = self.parameters.optimizations_per_minibatch._get(context)
> 12101 result = runner.run_learning(
12102 inputs=inputs,
12103 targets=targets,
12104 num_trials=num_trials,
12105 epochs=epochs,
12106 learning_rate=learning_rate,
12107 minibatch_size=minibatch_size,
12108 optimizations_per_minibatch=optimizations_per_minibatch,
12109 patience=patience,
12110 min_delta=min_delta,
12111 randomize_minibatches=randomize_minibatches,
12112 call_before_minibatch=call_before_minibatch,
12113 call_after_minibatch=call_after_minibatch,
12114 context=context,
12115 execution_mode=execution_mode,
12116 skip_initialization=skip_initialization,
12117 *args, **kwargs)
12119 context.remove_flag(ContextFlags.LEARNING_MODE)
12120 return result
File /opt/hostedtoolcache/Python/3.11.14/x64/lib/python3.11/site-packages/psyneulink/library/compositions/compositionrunner.py:465, in CompositionRunner.run_learning(self, inputs, targets, num_trials, epochs, learning_rate, minibatch_size, optimizations_per_minibatch, patience, min_delta, randomize_minibatches, synch_with_pnl_options, retain_in_pnl_options, call_before_minibatch, call_after_minibatch, context, execution_mode, skip_initialization, **kwargs)
462 run_trials = num_trials * stim_epoch if self._is_llvm_mode else None
464 # IMPLEMENTATION NOTE: for autodiff composition, the following executes a MINIBATCH's worth of training
--> 465 self._composition.run(inputs=minibatched_input,
466 num_trials=run_trials,
467 skip_initialization=skip_initialization,
468 skip_analyze_graph=True,
469 optimizations_per_minibatch=optimizations_per_minibatch,
470 synch_with_pnl_options=synch_with_pnl_options,
471 retain_in_pnl_options=retain_in_pnl_options,
472 execution_mode=execution_mode,
473 context=context,
474 **kwargs)
475 skip_initialization = True
477 if execution_mode is ExecutionMode.PyTorch:
File /opt/hostedtoolcache/Python/3.11.14/x64/lib/python3.11/site-packages/psyneulink/core/globals/context.py:747, in handle_external_context.<locals>.decorator.<locals>.wrapper(context, *args, **kwargs)
744 pass
746 try:
--> 747 return func(*args, context=context, **kwargs)
748 except TypeError as e:
749 # context parameter may be passed as a positional arg
750 if (
751 f"{func.__name__}() got multiple values for argument"
752 not in str(e)
753 ):
File /opt/hostedtoolcache/Python/3.11.14/x64/lib/python3.11/site-packages/psyneulink/library/compositions/autodiffcomposition.py:2033, in AutodiffComposition.run(self, synch_projection_matrices_with_torch, synch_node_variables_with_torch, synch_node_values_with_torch, synch_results_with_torch, retain_torch_trained_outputs, retain_torch_targets, retain_torch_losses, batched_results, context, *args, **kwargs)
2030 kwargs[RETAIN_IN_PNL_OPTIONS] = retain_in_pnl_options
2032 # Run AutodiffComposition
-> 2033 results = super(AutodiffComposition, self).run(*args, context=context, **kwargs)
2035 if EXECUTION_MODE in kwargs and kwargs[EXECUTION_MODE] is pnlvm.ExecutionMode.PyTorch:
2036 # Synchronize specified outcomes at end of run
2037 pytorch_rep = self.parameters.pytorch_representation.get(context)
File /opt/hostedtoolcache/Python/3.11.14/x64/lib/python3.11/site-packages/psyneulink/core/globals/context.py:747, in handle_external_context.<locals>.decorator.<locals>.wrapper(context, *args, **kwargs)
744 pass
746 try:
--> 747 return func(*args, context=context, **kwargs)
748 except TypeError as e:
749 # context parameter may be passed as a positional arg
750 if (
751 f"{func.__name__}() got multiple values for argument"
752 not in str(e)
753 ):
File /opt/hostedtoolcache/Python/3.11.14/x64/lib/python3.11/site-packages/psyneulink/core/compositions/composition.py:11808, in Composition.run(self, inputs, num_trials, initialize_cycle_values, reset_stateful_functions_to, reset_stateful_functions_when, skip_initialization, clamp_input, runtime_params, call_before_time_step, call_after_time_step, call_before_pass, call_after_pass, call_before_trial, call_after_trial, termination_processing, skip_analyze_graph, report_output, report_params, report_progress, report_simulations, report_to_devices, animate, log, scheduler, scheduling_mode, execution_mode, default_absolute_time_unit, context, base_context, **kwargs)
11804 execution_stimuli = None
11806 # execute processing, passing stimuli for this trial
11807 # IMPLEMENTATION NOTE: for autodiff, the following executes the forward pass for a single input
> 11808 trial_output = self.execute(inputs=execution_stimuli,
11809 scheduler=scheduler,
11810 termination_processing=termination_processing,
11811 call_before_time_step=call_before_time_step,
11812 call_before_pass=call_before_pass,
11813 call_after_time_step=call_after_time_step,
11814 call_after_pass=call_after_pass,
11815 reset_stateful_functions_to=reset_stateful_functions_to,
11816 context=context,
11817 base_context=base_context,
11818 clamp_input=clamp_input,
11819 runtime_params=runtime_params,
11820 skip_initialization=True,
11821 execution_mode=execution_mode,
11822 report=report,
11823 report_num=report_num,
11824 **kwargs
11825 )
11827 # ---------------------------------------------------------------------------------
11828 # store the result of this execution in case it will be the final result
11830 trial_output = copy_parameter_value(trial_output)
File /opt/hostedtoolcache/Python/3.11.14/x64/lib/python3.11/site-packages/psyneulink/core/globals/context.py:747, in handle_external_context.<locals>.decorator.<locals>.wrapper(context, *args, **kwargs)
744 pass
746 try:
--> 747 return func(*args, context=context, **kwargs)
748 except TypeError as e:
749 # context parameter may be passed as a positional arg
750 if (
751 f"{func.__name__}() got multiple values for argument"
752 not in str(e)
753 ):
File /opt/hostedtoolcache/Python/3.11.14/x64/lib/python3.11/site-packages/psyneulink/library/compositions/autodiffcomposition.py:1932, in AutodiffComposition.execute(self, inputs, num_trials, minibatch_size, optimizations_per_minibatch, do_logging, scheduler, termination_processing, call_before_minibatch, call_after_minibatch, call_before_time_step, call_before_pass, call_after_time_step, call_after_pass, reset_stateful_functions_to, context, base_context, clamp_input, targets, optimizer_params, runtime_params, execution_mode, skip_initialization, synch_with_pnl_options, retain_in_pnl_options, report_output, report_params, report_progress, report_simulations, report_to_devices, report, report_num)
1923 # Begin reporting of learning TRIAL:
1924 report(self,
1925 LEARN_REPORT,
1926 # EXECUTE_REPORT,
(...) 1929 content='trial_start',
1930 context=context)
-> 1932 self._build_pytorch_representation(optimizer_params=optimizer_params,
1933 learning_rate=self.parameters.learning_rate.get(context),
1934 context=context, base_context=base_context)
1935 trained_output_values, all_output_values = \
1936 self.autodiff_forward(inputs=autodiff_inputs,
1937 targets=autodiff_targets,
(...) 1941 scheduler=scheduler,
1942 context=context)
1943 execution_phase = context.execution_phase
File /opt/hostedtoolcache/Python/3.11.14/x64/lib/python3.11/site-packages/psyneulink/core/globals/context.py:747, in handle_external_context.<locals>.decorator.<locals>.wrapper(context, *args, **kwargs)
744 pass
746 try:
--> 747 return func(*args, context=context, **kwargs)
748 except TypeError as e:
749 # context parameter may be passed as a positional arg
750 if (
751 f"{func.__name__}() got multiple values for argument"
752 not in str(e)
753 ):
File /opt/hostedtoolcache/Python/3.11.14/x64/lib/python3.11/site-packages/psyneulink/library/compositions/autodiffcomposition.py:1267, in AutodiffComposition._build_pytorch_representation(self, learning_rate, optimizer_params, context, new, base_context)
1264 pass
1265 else:
1266 # Otherwise, just update it
-> 1267 pytorch_rep._update_optimizer_params(old_opt,
1268 optimizer_params,
1269 Context(source=ContextFlags.METHOD,
1270 runmode=context.runmode,
1271 execution_id=context.execution_id))
1272 # Set up loss function
1273 if self.loss_function is not None:
File /opt/hostedtoolcache/Python/3.11.14/x64/lib/python3.11/site-packages/psyneulink/library/compositions/pytorchwrappers.py:844, in PytorchCompositionWrapper._update_optimizer_params(self, optimizer, optimizer_params_user_specs, context)
839 if source == CONSTRUCTOR and self.optimizer:
840 # If user has specified dict with learning_rates in call to _build_pytorch_representation,
841 # need to update the construct_param_groups with specififed values
842 self._update_constructor_param_groups(self.composition, optimizer_params_user_specs)
--> 844 self._assign_learning_rates(optimizer,
845 optimizer_params_user_parsed,
846 optimizer_torch_params_full_with_specified,
847 run_time_default_learning_rate,
848 source,
849 context)
File /opt/hostedtoolcache/Python/3.11.14/x64/lib/python3.11/site-packages/psyneulink/library/compositions/pytorchwrappers.py:1031, in PytorchCompositionWrapper._assign_learning_rates(self, optimizer, optimizer_params_user_parsed, optimizer_torch_params_full_with_specified, run_time_default_learning_rate, source, context)
1029 default_learning_rate = old_param_group['lr']
1030 for param in old_param_group['params']:
-> 1031 projection = self._torch_params_to_projections(old_param_groups)[param]
1032 specified_learning_rate = (
1033 self._get_specified_learning_rate_for_param(param, projection,
1034 optimizer_params_user_parsed,
1035 run_time_default_learning_rate,
1036 source, context))
1037 if specified_learning_rate is not False:
File /opt/hostedtoolcache/Python/3.11.14/x64/lib/python3.11/site-packages/psyneulink/library/compositions/pytorchwrappers.py:1304, in PytorchCompositionWrapper._torch_params_to_projections(self, param_groups)
1302 # Give subclasses a chance for custom handling of param->projection mapping
1303 for comp_wrapper in self.get_all_nested_composition_wrappers():
-> 1304 torch_params_to_projections.update(comp_wrapper._torch_params_to_projections(param_groups))
1305 return torch_params_to_projections
File /opt/hostedtoolcache/Python/3.11.14/x64/lib/python3.11/site-packages/psyneulink/library/compositions/pytorchwrappers.py:1301, in PytorchCompositionWrapper._torch_params_to_projections(self, param_groups)
1299 for proj in self.wrapped_projections:
1300 if proj.name in self._pnl_refs_to_torch_param_names:
-> 1301 torch_params_to_projections.update({self.get_torch_param_for_projection(proj): proj})
1302 # Give subclasses a chance for custom handling of param->projection mapping
1303 for comp_wrapper in self.get_all_nested_composition_wrappers():
File /opt/hostedtoolcache/Python/3.11.14/x64/lib/python3.11/site-packages/psyneulink/library/compositions/pytorchwrappers.py:1250, in PytorchCompositionWrapper.get_torch_param_for_projection(self, projection)
1248 projection_name = projection.name if isinstance(projection, Projection) else projection
1249 param_name = self._pnl_refs_to_torch_param_names[projection_name].param_name
-> 1250 torch_long_param_name = self._torch_param_short_to_long_names_map[param_name]
1251 for param_tuple in self.named_parameters():
1252 # param_tuple is a tuple of (name, torch.nn.Parameter)
1253 if torch_long_param_name == param_tuple[0]:
File /opt/hostedtoolcache/Python/3.11.14/x64/lib/python3.11/site-packages/psyneulink/library/compositions/pytorchwrappers.py:1312, in PytorchCompositionWrapper._torch_param_short_to_long_names_map(self)
1307 @property
1308 def _torch_param_short_to_long_names_map(self)->dict:
1309 """Return map of short torch Parameter names to their full (hierarchical) names in named_parameters()
1310 The "full" names should include prefixes for parameters in nested PytorchCompositionWrappers.
1311 """
-> 1312 return {k.split('.')[-1]:k for k in [p[0] for p in self.named_parameters()]}
File /opt/hostedtoolcache/Python/3.11.14/x64/lib/python3.11/site-packages/psyneulink/library/compositions/pytorchwrappers.py:1312, in <listcomp>(.0)
1307 @property
1308 def _torch_param_short_to_long_names_map(self)->dict:
1309 """Return map of short torch Parameter names to their full (hierarchical) names in named_parameters()
1310 The "full" names should include prefixes for parameters in nested PytorchCompositionWrappers.
1311 """
-> 1312 return {k.split('.')[-1]:k for k in [p[0] for p in self.named_parameters()]}
File /opt/hostedtoolcache/Python/3.11.14/x64/lib/python3.11/site-packages/torch/nn/modules/module.py:2706, in Module.named_parameters(self, prefix, recurse, remove_duplicate)
2679 r"""Return an iterator over module parameters, yielding both the name of the parameter as well as the parameter itself.
2680
2681 Args:
(...) 2698
2699 """
2700 gen = self._named_members(
2701 lambda module: module._parameters.items(),
2702 prefix=prefix,
2703 recurse=recurse,
2704 remove_duplicate=remove_duplicate,
2705 )
-> 2706 yield from gen
File /opt/hostedtoolcache/Python/3.11.14/x64/lib/python3.11/site-packages/torch/nn/modules/module.py:2641, in Module._named_members(self, get_members_fn, prefix, recurse, remove_duplicate)
2635 memo = set()
2636 modules = (
2637 self.named_modules(prefix=prefix, remove_duplicate=remove_duplicate)
2638 if recurse
2639 else [(prefix, self)]
2640 )
-> 2641 for module_prefix, module in modules:
2642 members = get_members_fn(module)
2643 for k, v in members:
File /opt/hostedtoolcache/Python/3.11.14/x64/lib/python3.11/site-packages/torch/nn/modules/module.py:2863, in Module.named_modules(self, memo, prefix, remove_duplicate)
2861 continue
2862 submodule_prefix = prefix + ("." if prefix else "") + name
-> 2863 yield from module.named_modules(
2864 memo, submodule_prefix, remove_duplicate
2865 )
File /opt/hostedtoolcache/Python/3.11.14/x64/lib/python3.11/site-packages/torch/nn/modules/module.py:2863, in Module.named_modules(self, memo, prefix, remove_duplicate)
2861 continue
2862 submodule_prefix = prefix + ("." if prefix else "") + name
-> 2863 yield from module.named_modules(
2864 memo, submodule_prefix, remove_duplicate
2865 )
File /opt/hostedtoolcache/Python/3.11.14/x64/lib/python3.11/site-packages/torch/nn/modules/module.py:2863, in Module.named_modules(self, memo, prefix, remove_duplicate)
2861 continue
2862 submodule_prefix = prefix + ("." if prefix else "") + name
-> 2863 yield from module.named_modules(
2864 memo, submodule_prefix, remove_duplicate
2865 )
File /opt/hostedtoolcache/Python/3.11.14/x64/lib/python3.11/site-packages/torch/nn/modules/module.py:2818, in Module.named_modules(self, memo, prefix, remove_duplicate)
2815 for _, module in self.named_modules():
2816 yield module
-> 2818 def named_modules(
2819 self,
2820 memo: Optional[set["Module"]] = None,
2821 prefix: str = "",
2822 remove_duplicate: bool = True,
2823 ):
2824 r"""Return an iterator over all modules in the network, yielding both the name of the module as well as the module itself.
2825
2826 Args:
(...) 2851
2852 """
2853 if memo is None:
KeyboardInterrupt:
import matplotlib.pyplot as plt
import numpy as np
TOTAL_NUM_STIMS = len(trials)
TARGETS = np.array(trials[1:] + [one_hot_encode(0, 11)])
curriculum_type = BLOCKED
fig, axes = plt.subplots(1, 1, figsize=(12, 5))
# L1 of loss
axes.plot((np.abs(ego_model.results[1:TOTAL_NUM_STIMS, 2] - TARGETS[:TOTAL_NUM_STIMS - 1])).sum(-1))
axes.set_xlabel('Stimuli')
axes.set_ylabel('Loss')
plt.suptitle(f"{curriculum_type} Training")
plt.show()
Run the model for the interleaved paradigm. What do you expect? Compare the two results and explain the differences.