9.1 ISC-CI Model#
The ISC-CI model introducing a mechanism for context inference, based on the key assumption that temporal co-occurrence provides a useful basis for inferring shared context. Specifically, it assumes that (1) objects occurring together in a given context tend to share the properties elicited by that context; (2) these co-occurrence statistics are learned over the course of development; and (3) this implicit knowledge provides a basis for inferring, from a few examples of objects encountered in a new context, both which features are relevant in that context and what other objects are likely to occur in that context.
To make these ideas clear, consider the contexts in which you might encounter different kinds of birds: a bird-watching field trip in science class, a visit to the bird section of the zoo, and a picture book about birds. Each situation involves multiple types of birds (e.g., robins, crows, and ravens) and exposure to multiple bird-related properties (e.g., can-fly, eats-worms, is-bird) in various combinations. After these experiences, encountering a new context in which birds are relevant (e.g., learning that crows and ravens have hollow bones in the bird section of the Natural History museum) is likely to be interpreted as relating specifically to birds and their properties, implying that other birds like robins may also occur in this new context, and that they will share similar properties (e.g., robins also have hollow bones).
Conversely, contexts such as a science lesson on aerodynamics, a visit to a flight exhibit at a science museum, and a film on the history of flight are likely to involve multiple types of flying things (e.g., crows, airplanes, and butterflies) and flight-related properties (e.g., can-fly, has-wings, seen-in-the-sky). This suggests that a new context involving flying objects such as crows and airplanes (e.g., learning that crows and airplanes are associated with Bernoulli’s principle) likely relates to all things that can fly, implying that other flying things like butterflies may also occur in this new context and, again, share similar properties (e.g., butterflies are also associated with Bernoulli’s principle).
Thus, the properties shared by items encountered in a situation can provide a clue about what the current context is, what properties are currently important, and what other items are likely or unlikely also to be observed. The central hypothesis embodied by the ISC-CI model is that learning such environmental structure can support future inferences about which features might be relevant in novel contexts, based on the distribution of items that co-occur in those contexts. That is, observing that a new context involves a certain set of objects (e.g., both robins and airplanes) provides evidence that certain features will be context-relevant (e.g., can-fly and has-wings), but not others (e.g., lays-eggs), based on past experience.
Importantly, this process is graded and probabilistic rather than absolute, as any given set of objects can co-occur in different contexts at different frequencies. In particular, features that are broadly true of many objects are less likely to be relevant in a new context than features that are true of the more limited set of objects seen in that context (Griffiths et al., 2010; Xu & Tenenbaum, 2007). This is because there is a low likelihood of observing any particular set of objects in a broad context: there are many animals, but few Corvidae, so it is more likely that a context involving both crows and ravens relates to Corvidae specifically than it is that this context relates to animals in general. This is because the probability of observing both crows and ravens in the context of animals is lower than the probability of oberseving both crows and ravens in the context of Corvidae.
Setup and Installation:
%%capture
%pip install psyneulink
import psyneulink as pnl
import pandas as pd
import random
Generating the Training Data#
Feature Co-occurrences#
We design a training environment that simulates experiencing object co-occurrences throughout learning under the key assumption noted just above. The environment consisted of a series of episodes corresponding to different contexts. Each context involved a set of objects that share a common semantic feature (e.g., things that are birds, things that can fly, things that are found in the zoo, etc.), with each feature represented by a single output unit as implemented by the feature labels in the ISC-CI model.
We generate the episodes using the objects and features in the Leuven Concepts Database (De Deyne & Storms, 2008; Storms, 2001; Ruts et al., 2004). That database contains a matrix of binary judgments provided by human raters indicating, for each object-feature pairing, whether the object possess the feature (e.g., does a bear weigh more than 100lbs? Are kangaroos found in zoos?).
Let’s explore the dataset
FEATURE_PATH = 'https://raw.githubusercontent.com/PrincetonUniversity/NEU-PSY-502/refs/heads/main/data/isc_ci/features.csv'
feature_df = pd.read_csv(FEATURE_PATH, index_col=0)
feature_df.head()
| is small | is a bird | is an animal | is big | can fly | is an insect | mammal | is a fish | lays eggs | is brown | ... | is for all ages | you can play different notes whit it | can be used to put something in | can be bought in sports store | costs a lot of money | drives above the ground | driven by 1 person | used in water | used in the house | worn often | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| monkey | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| beaver | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| bison | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| dromedary | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | 0 | 1 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| squirrel | 1 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
5 rows × 385 columns
As stated above, a context is defined by the common feature shared by the objects in that context. For this dataset, which objects might occur in the “eats mice” context?
✅ Solution
We can find the objects that might occur in the “eats mice” context by filtering the dataset for the feature “eats mice” and extracting the corresponding object names:
eats_mice = feature_df[feature_df['eats mice'] == 1].index.tolist()
As stated above, encountering different objects in the same episode provides evidence to the agent that they are in a specific context. For this dataset, which context might be defined by the object “owl” (and which by “falcon”)?
✅ Solution
The contexts are defined by the features of the objects:
contexts_owl = feature_df.columns[feature_df.loc['owl'] == 1]
contexts_falcon = feature_df.columns[feature_df.loc['falcon'] == 1]
print('Contexts defined by "owl":', contexts_owl)
print('Contexts defined by "falcon":', contexts_falcon)
Objects can appear in various contexts. So for two given objects, they might define multiple contexts. What possible contexts are defined by “owl” and “falcon” together?
✅ Solution
The possible contexts are defined by the features that are shared by “owl” and “falcon”:
contexts_both = feature_df.columns[(feature_df.loc['owl'] == 1) & (feature_df.loc['falcon'] == 1)]
print(contexts_both)
Now (using the theory from above), try to answer the following question: Given an agent experiences an episode with “owl” and “falcon” defining the context. Would the agent be more surprised by encountering a “cat” in this context or “penguin”?
(This is not a straight forward question, and you are not supposed to give a definite answer. Think about different (probabilistic/statistical) features of the world influencing the agent’s prediction).
Tip: Here we make the (unreasonable) assumption that contexts are uniformly distributed. In other words that any episode (being on a bird-watching tour or being in a science lesson) has occurred equally often in the past.
💡 Hint a
In the exercise above, we’ve seen that “owl” and “falcon” can elicit different contexts. But is there a way to “quantify” which of these contexts is more likely to be elicited?
Tip: This depends on how likely it is to encounter both an “owl” and a “falcon” in any of the given contexts.
💡 Hint b
We calculate the probabilities from above:
contexts_both = feature_df.columns[(feature_df.loc['owl'] == 1) & (feature_df.loc['falcon'] == 1)]
context_probabilities = {}
for c in contexts_both:
objects = feature_df[feature_df[c] == 1].index.tolist()
nr_objects = len(objects)
# Simplification (objects are equally likely):
# The probability for encountering any object is 1 / nr_objects
# => the probability of 'drawing' two objects is 1 / nr_objects^2
probability = 1 / nr_objects**2
context_probabilities[c] = probability
print(context_probabilities)
print()
most_probabl_context = max(context_probabilities, key=context_probabilities.get)
print(most_probabl_context)
💡 Hint c
The code above shows that the most probable context is “eats mice”. But “penguins” don’t eat mice while “cats” do (You can convince yourself by querying the database :). So if the agent encounters a “cat” it should be less surprised than if it encounters a “penguin”.
However, this is just the most probable context. Although “penguin” doesn’t appear in the most probable context of “owl” and “falcon”, this might be offset by “penguin” appearing in more probable contexts than “cat”.
✅ Solution
First, we calculate the probabilities of encountering “owl” and “falcon” in any given context. We normalize these probabilties and use them as weights: We add all the probabilities of contexts where “penguin” also appears vs “cat”:
contexts_both = feature_df.columns[(feature_df.loc['owl'] == 1) & (feature_df.loc['falcon'] == 1)]
context_probabilities = {}
for c in contexts_both:
objects = feature_df[feature_df[c] == 1].index.tolist()
nr_objects = len(objects)
probability = 1 / nr_objects**2
context_probabilities[c] = probability
# normalize the to get the probabilty of a certain context beeing evoked
sum_probs = sum(context_probabilities.values())
normalized = {c: p / sum_probs for c, p in context_probabilities.items()}
penguin_weight = 0
penguin_nr = 0
cat_weight = 0
cat_nr = 0
for c, v in normalized.items():
objects = feature_df[feature_df[c] == 1].index.tolist()
# the context evokes penguin
if 'penguin' in objects:
penguin_weight += v
penguin_nr += 1
# the context evokes cat
if 'cat' in objects:
cat_weight += v
cat_nr += 1
print(f'penguins({penguin_nr}): {penguin_weight}')
print(f'cats({cat_nr}): {cat_weight}')
Embedding#
The ISC-CI model uses as input a word embedding (not a one-hot encoding). This embedding can be interpreted as “context independent” representation. Here, we load the embeddings:
EMBEDDING_PATH = 'https://raw.githubusercontent.com/PrincetonUniversity/NEU-PSY-502/refs/heads/main/data/isc_ci/embeddings.csv'
embeddings_df = pd.read_csv(EMBEDDING_PATH, index_col=0)
chicken_embedding = embeddings_df.loc['chicken']
chicken_embedding
0 0.119731
1 0.987248
2 0.982023
3 0.410961
4 0.451753
...
59 0.022675
60 0.326714
61 0.014851
62 0.035483
63 0.982558
Name: chicken, Length: 64, dtype: float64
We can ask the same question as above: Can you think about a way of using the embeddings of “owl”, “falcon”, “cat” and “penguin” to quantify weather the agent would be more surprised by encountering a “cat” or a “penguin” in the context of “owl” and “falcon”?
💡 Hint
With word embeddings, we can calculate similarities between words by using the distance.
✅ Solution
We calculate the distance between the embeddings of “owl” and “falcon” to the embeddings of “cat” and “penguin”:
owl_emb = embeddings_df.loc['owl']
falcon_emb = embeddings_df.loc['falcon']
cat_emb = embeddings_df.loc['cat']
penguin_emb = embeddings_df.loc['penguin']
owl_falcon_emb = (owl_emb + falcon_emb) / 2
cat_distance = ((owl_falcon_emb - cat_emb)**2).sum()
penguin_distance = ((owl_falcon_emb - penguin_emb)**2).sum()
print('cat distance ', cat_distance)
print('penguin distance ', penguin_distance)
Note, the “context”-calculations and “embedding”-calculations lead to different predictions:
context -> “cat” is less surprising in “owl”, “falcon” context
embedding -> “penguin” is less surprising in “owl”, “falcon” context
Training Data#
Now, we construct a set of episodes by uniformly sampling from the set of features with replacement, so that each episode involved one shared semantic feature that defined the associated context. Given this feature, we generated a support set by uniformly sampling two items sharing the feature, and a query set by uniformly sampling one additional item sharing the feature (positive) and one that is not sharing the feature (negative). Note, that as above-mentioned, any given set of objects can co-occur in multiple contexts (in other words can share multiple features), however, for features tha are broadly true of many objects, it is less likely for each specific pair of objects to be sampled. For example, if the feature is “eats mice”, the likelihood of sampling exactly {“owl”, “falcon”} is higher than if the feature is “is a bird” (since there are many more birds than mice eaters).
def get_random_episode():
# randomly pick a context (feature)
feature = random.choice(feature_df.columns)
# one hot encoded feature vector is our context:
context_label = [0] * len(feature_df.columns)
context_label[feature_df.columns.get_loc(feature)] = 1
# get objects that have this feature by name
objects_included = feature_df[feature_df[feature] == 1].index.tolist()
# get objects that don't have this feature
objects_excluded = feature_df[feature_df[feature] == 0].index.tolist()
# randomly pick two supports (can be the same twice)
support = random.choices(objects_included, k=2)
# the support vector is the embedding of the two supports
support_vector_1 = embeddings_df.loc[support[0]]
support_vector_2 = embeddings_df.loc[support[1]]
# decide weather to pick an object that is in the context or not
choice = random.choice([0, 1])
if choice == 0:
# pick an object that is not in the context
query = random.choice(objects_excluded)
else:
# pick an object that is not in the context
query = random.choice(objects_included)
# the query vector is the embedding of the query
query_vector = embeddings_df.loc[query]
return {
'context_label': context_label,
'support_1': support_vector_1,
'support_2': support_vector_2,
'query': query_vector,
'response': [1, 0] if choice > .5 else [0, 1]
}
PsyNeuLink Model#
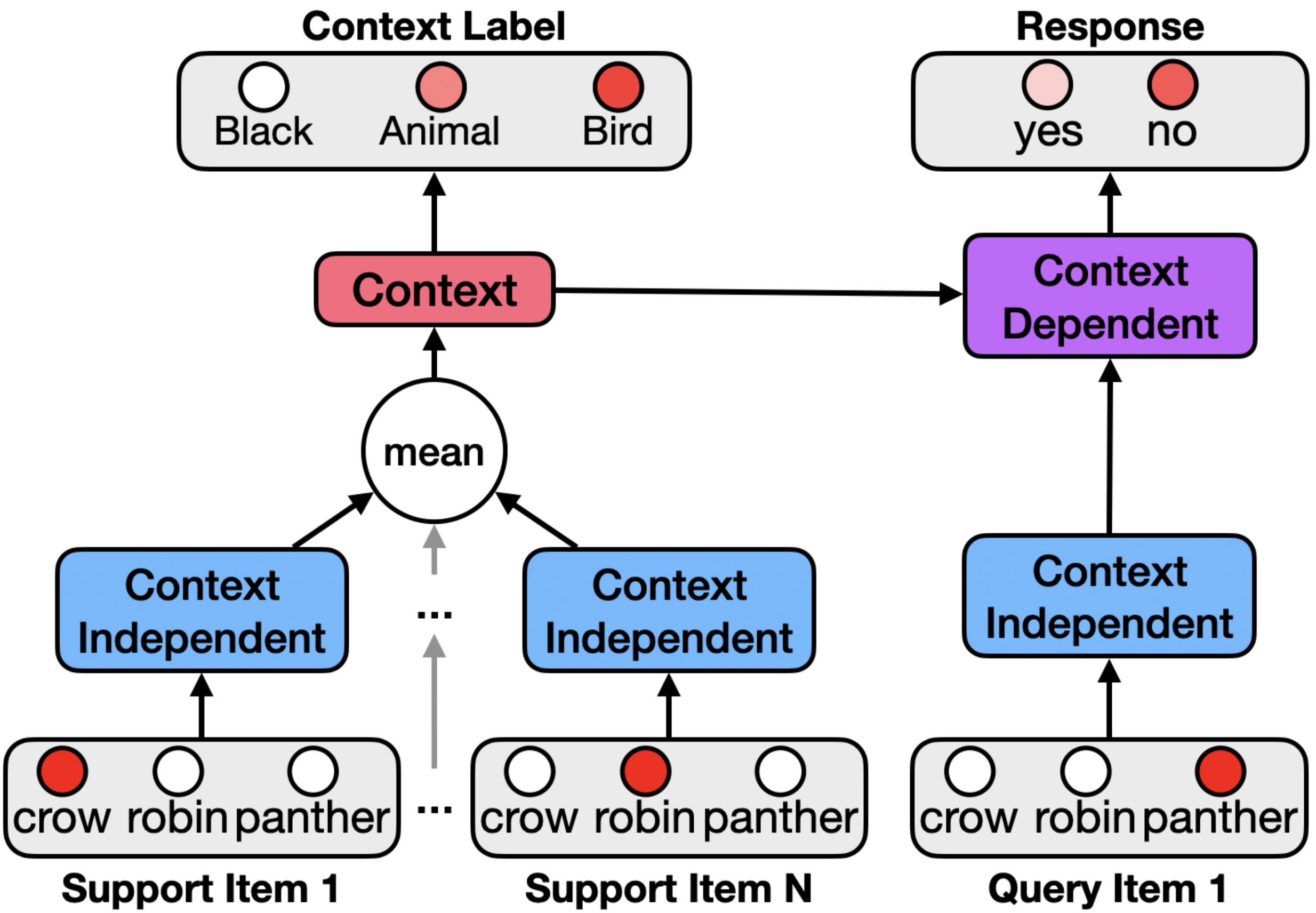
We design the model to infer the context from the objects in the support set and use this to make predictions about objects in the query set. The model’s architecture contains a context independent layer that encodes cross-context information, a context layer that encodes information about which features are relevant in the given context, and a context-dependent layer which selectively encodes context-relevant information. The ISC-CI model processes the items in the support set and uses this both to generate an internal representation of context and provide a predicted context-specific shared semantic feature label as output.
The ISC-CI model makes inferences about the current context using objects in the support set by sequentially observing each object in the support set, encoding each in the context independent layer, and integrating these representations by taking their average. This can be viewed as a very simple form of recurrence that accumulates (by linearly integrating and normalizing) activity across the items in the support set in the context independent layer.
The ISC-CI model makes predictions about which items in the query set occur in the current context. It does so by forming a context dependent representation that takes into account the context inferred from the support set together with the context-independent representation of the query set item. The model then uses this context dependent representation to activate the binary yes/no output unit indicating the likelihood that the query set item occurs in the current context.
Convince yourself that as context independent layer, we can use the embeddings of the support and query labels. In our PsyNeuLink implementation, we will “skip” the one-hot encoded labels and use the embeddings loaded earlier as input directly.
✅ Solution
The context independent layer captures context independent statistical properties of the objects (words). Word embeddings are a good representation of these properties and in many cases specifically trained to capture these properties (compare, for example, word2vec).
Explain -in your own words - why calculating the mean of the support embeddings a form of integration. Could you think about a different way of integrating the support embeddings?
💡 Hint
The mean of the support embeddings combines the information of multiple support vectors into one representation. However, does the order of the support vectors matter? Could you think about a way of integrating the support embeddings in a way that most recent support vectors have more weight than older ones?
✅ Solution
The mean of the support embeddings is a form of integration because it combines the information from multiple support embeddings into a single representation. This allows the model to capture the shared properties of the objects. Other ways of integrating the support embeddings could include using a weighted sum or recurrent neural networks (RNNs or other forms of temporal integration like LSTM or GRU). These methods would allow to integrate the support vectors “over time” and take into account the order of the support embeddings.
Before implementing the model, think about the dimensions of the different layers:
context independent, mean, context, context label, context dependent, and response
✅ Solution
The dimensions of the different layers are as follows:
context independent: embedding dimension
mean: embedding dimension
context: arbitrary dimension (128 in the paper)
context label: number of features in the feature set
context dependent: arbitrary but the same as the context dimension
response: 2 (yes/no)
N_EMBEDDING = embeddings_df.shape[1]
HIDDEN_CONTEXT = 128
N_CONTEXT = feature_df.shape[1]
LEARNING_RATE = 0.01
def create_model(
):
## Mechanisms
# Here, we use 2 support vectors (but we could use more)
support_1 = pnl.TransferMechanism(
name='support_1',
input_shapes=N_EMBEDDING
)
support_2 = pnl.TransferMechanism(
name='support_2',
input_shapes=N_EMBEDDING
)
# The mean of the support vectors is calculated by adding them
# and multiplying by .5
mean = pnl.TransferMechanism(
name='mean',
input_shapes=N_EMBEDDING,
function=pnl.Linear(slope=.5),
)
context = pnl.TransferMechanism(
name='context',
input_shapes=HIDDEN_CONTEXT,
function=pnl.ReLU()
)
context_label = pnl.TransferMechanism(
name='context_label',
input_shapes=N_CONTEXT,
function=pnl.SoftMax()
)
query = pnl.TransferMechanism(
name='query',
input_shapes=N_EMBEDDING
)
context_dependent = pnl.TransferMechanism(
name='context_dependent',
input_shapes=HIDDEN_CONTEXT,
function=pnl.ReLU()
)
response = pnl.TransferMechanism(
name='response',
input_shapes=2,
function=pnl.SoftMax(),
)
## Projections
projection_mean_to_context = pnl.MappingProjection(
name='projection_mean_to_context',
matrix=pnl.RandomMatrix(0, .01),
learnable=True
)
projection_query_to_context_dependent = pnl.MappingProjection(
name='projection_query_to_context_dependent',
matrix=pnl.RandomMatrix(0, .01),
learnable=True
)
projection_context_to_context_label = pnl.MappingProjection(
name='projection_context_dependent_to_context_out',
matrix=pnl.RandomMatrix(0, .01),
learnable=True
)
projection_context_dependent_to_response = pnl.MappingProjection(
name='projection_query_dependent_to_response',
matrix=pnl.RandomMatrix(0, .01),
learnable=True
)
## Pathways
model = pnl.AutodiffComposition(
name='embedding',
pathways=[
[support_1, pnl.IDENTITY_MATRIX, mean],
[support_2, pnl.IDENTITY_MATRIX, mean],
[query, projection_query_to_context_dependent, context_dependent],
[mean, projection_mean_to_context, context],
[context, pnl.IDENTITY_MATRIX, context_dependent],
[context, projection_context_to_context_label, context_label],
[context_dependent, projection_context_dependent_to_response, response]
],
learning_rate=LEARNING_RATE
)
return (model, query, support_1, support_2,
context_label, response)
isc_ci_model, query, support_1, support_2, context_label, response = create_model()
isc_ci_model.show_graph(output_fmt='jupyter')
---------------------------------------------------------------------------
FileNotFoundError Traceback (most recent call last)
File /opt/hostedtoolcache/Python/3.11.14/x64/lib/python3.11/site-packages/graphviz/backend/execute.py:76, in run_check(cmd, input_lines, encoding, quiet, **kwargs)
75 kwargs['stdout'] = kwargs['stderr'] = subprocess.PIPE
---> 76 proc = _run_input_lines(cmd, input_lines, kwargs=kwargs)
77 else:
File /opt/hostedtoolcache/Python/3.11.14/x64/lib/python3.11/site-packages/graphviz/backend/execute.py:96, in _run_input_lines(cmd, input_lines, kwargs)
95 def _run_input_lines(cmd, input_lines, *, kwargs):
---> 96 popen = subprocess.Popen(cmd, stdin=subprocess.PIPE, **kwargs)
98 stdin_write = popen.stdin.write
File /opt/hostedtoolcache/Python/3.11.14/x64/lib/python3.11/subprocess.py:1026, in Popen.__init__(self, args, bufsize, executable, stdin, stdout, stderr, preexec_fn, close_fds, shell, cwd, env, universal_newlines, startupinfo, creationflags, restore_signals, start_new_session, pass_fds, user, group, extra_groups, encoding, errors, text, umask, pipesize, process_group)
1023 self.stderr = io.TextIOWrapper(self.stderr,
1024 encoding=encoding, errors=errors)
-> 1026 self._execute_child(args, executable, preexec_fn, close_fds,
1027 pass_fds, cwd, env,
1028 startupinfo, creationflags, shell,
1029 p2cread, p2cwrite,
1030 c2pread, c2pwrite,
1031 errread, errwrite,
1032 restore_signals,
1033 gid, gids, uid, umask,
1034 start_new_session, process_group)
1035 except:
1036 # Cleanup if the child failed starting.
File /opt/hostedtoolcache/Python/3.11.14/x64/lib/python3.11/subprocess.py:1955, in Popen._execute_child(self, args, executable, preexec_fn, close_fds, pass_fds, cwd, env, startupinfo, creationflags, shell, p2cread, p2cwrite, c2pread, c2pwrite, errread, errwrite, restore_signals, gid, gids, uid, umask, start_new_session, process_group)
1954 if err_filename is not None:
-> 1955 raise child_exception_type(errno_num, err_msg, err_filename)
1956 else:
FileNotFoundError: [Errno 2] No such file or directory: PosixPath('dot')
The above exception was the direct cause of the following exception:
ExecutableNotFound Traceback (most recent call last)
File /opt/hostedtoolcache/Python/3.11.14/x64/lib/python3.11/site-packages/IPython/core/formatters.py:1036, in MimeBundleFormatter.__call__(self, obj, include, exclude)
1033 method = get_real_method(obj, self.print_method)
1035 if method is not None:
-> 1036 return method(include=include, exclude=exclude)
1037 return None
1038 else:
File /opt/hostedtoolcache/Python/3.11.14/x64/lib/python3.11/site-packages/graphviz/jupyter_integration.py:98, in JupyterIntegration._repr_mimebundle_(self, include, exclude, **_)
96 include = set(include) if include is not None else {self._jupyter_mimetype}
97 include -= set(exclude or [])
---> 98 return {mimetype: getattr(self, method_name)()
99 for mimetype, method_name in MIME_TYPES.items()
100 if mimetype in include}
File /opt/hostedtoolcache/Python/3.11.14/x64/lib/python3.11/site-packages/graphviz/jupyter_integration.py:98, in <dictcomp>(.0)
96 include = set(include) if include is not None else {self._jupyter_mimetype}
97 include -= set(exclude or [])
---> 98 return {mimetype: getattr(self, method_name)()
99 for mimetype, method_name in MIME_TYPES.items()
100 if mimetype in include}
File /opt/hostedtoolcache/Python/3.11.14/x64/lib/python3.11/site-packages/graphviz/jupyter_integration.py:112, in JupyterIntegration._repr_image_svg_xml(self)
110 def _repr_image_svg_xml(self) -> str:
111 """Return the rendered graph as SVG string."""
--> 112 return self.pipe(format='svg', encoding=SVG_ENCODING)
File /opt/hostedtoolcache/Python/3.11.14/x64/lib/python3.11/site-packages/graphviz/piping.py:104, in Pipe.pipe(self, format, renderer, formatter, neato_no_op, quiet, engine, encoding)
55 def pipe(self,
56 format: typing.Optional[str] = None,
57 renderer: typing.Optional[str] = None,
(...) 61 engine: typing.Optional[str] = None,
62 encoding: typing.Optional[str] = None) -> typing.Union[bytes, str]:
63 """Return the source piped through the Graphviz layout command.
64
65 Args:
(...) 102 '<?xml version='
103 """
--> 104 return self._pipe_legacy(format,
105 renderer=renderer,
106 formatter=formatter,
107 neato_no_op=neato_no_op,
108 quiet=quiet,
109 engine=engine,
110 encoding=encoding)
File /opt/hostedtoolcache/Python/3.11.14/x64/lib/python3.11/site-packages/graphviz/_tools.py:185, in deprecate_positional_args.<locals>.decorator.<locals>.wrapper(*args, **kwargs)
177 wanted = ', '.join(f'{name}={value!r}'
178 for name, value in deprecated.items())
179 warnings.warn(f'The signature of {func_name} will be reduced'
180 f' to {supported_number} positional arg{s_}{qualification}'
181 f' {list(supported)}: pass {wanted} as keyword arg{s_}',
182 stacklevel=stacklevel,
183 category=category)
--> 185 return func(*args, **kwargs)
File /opt/hostedtoolcache/Python/3.11.14/x64/lib/python3.11/site-packages/graphviz/piping.py:121, in Pipe._pipe_legacy(self, format, renderer, formatter, neato_no_op, quiet, engine, encoding)
112 @_tools.deprecate_positional_args(supported_number=1, ignore_arg='self')
113 def _pipe_legacy(self,
114 format: typing.Optional[str] = None,
(...) 119 engine: typing.Optional[str] = None,
120 encoding: typing.Optional[str] = None) -> typing.Union[bytes, str]:
--> 121 return self._pipe_future(format,
122 renderer=renderer,
123 formatter=formatter,
124 neato_no_op=neato_no_op,
125 quiet=quiet,
126 engine=engine,
127 encoding=encoding)
File /opt/hostedtoolcache/Python/3.11.14/x64/lib/python3.11/site-packages/graphviz/piping.py:149, in Pipe._pipe_future(self, format, renderer, formatter, neato_no_op, quiet, engine, encoding)
146 if encoding is not None:
147 if codecs.lookup(encoding) is codecs.lookup(self.encoding):
148 # common case: both stdin and stdout need the same encoding
--> 149 return self._pipe_lines_string(*args, encoding=encoding, **kwargs)
150 try:
151 raw = self._pipe_lines(*args, input_encoding=self.encoding, **kwargs)
File /opt/hostedtoolcache/Python/3.11.14/x64/lib/python3.11/site-packages/graphviz/backend/piping.py:212, in pipe_lines_string(engine, format, input_lines, encoding, renderer, formatter, neato_no_op, quiet)
206 cmd = dot_command.command(engine, format,
207 renderer=renderer,
208 formatter=formatter,
209 neato_no_op=neato_no_op)
210 kwargs = {'input_lines': input_lines, 'encoding': encoding}
--> 212 proc = execute.run_check(cmd, capture_output=True, quiet=quiet, **kwargs)
213 return proc.stdout
File /opt/hostedtoolcache/Python/3.11.14/x64/lib/python3.11/site-packages/graphviz/backend/execute.py:81, in run_check(cmd, input_lines, encoding, quiet, **kwargs)
79 except OSError as e:
80 if e.errno == errno.ENOENT:
---> 81 raise ExecutableNotFound(cmd) from e
82 raise
84 if not quiet and proc.stderr:
ExecutableNotFound: failed to execute PosixPath('dot'), make sure the Graphviz executables are on your systems' PATH
<graphviz.graphs.Digraph at 0x7fce8162c910>
The activation function of the context label and the response is SoftMax. Do you think these are reasonable activation functions? If so, why?
✅ Solution
The SoftMax activation function is reasonable for the context label because it outputs a probability distribution over the features. In the section above (generating the training data), we discussed that a context is exactly defined by one feature (a feature that is shared by all support items).
The SoftMax activation function is also reasonable for the response for the same reason. The response is a binary classification task (yes/no) and the SoftMax activation function captures this.
Training Loop#
The model is now trained on 20,000 episodes.
Note, that this is not enough for the model to converge and just illustrates how a training loop would look like. We don’t expect reasonable predictions after training.
tot_reps = 1
examples_per_rep = 20_000
n_epochs = 1
for reps in range(tot_reps):
inputs_dict = {
query: [],
support_1: [],
support_2: [],
}
targets_dict = {
context_label: [],
response: [],
}
# Get Trainign
i = 0
while i < examples_per_rep:
example = get_random_episode()
# Append the input to the dictionary
inputs_dict[query].append(example['query'])
inputs_dict[support_1].append(example['support_1'])
inputs_dict[support_2].append(example['support_2'])
# Append the targets to the dictionary
targets_dict[context_label].append(example['context_label'])
targets_dict[response].append(example['response'])
i += 1
# Train the network for `n_epochs`
result = isc_ci_model.learn(
inputs={
'inputs': inputs_dict,
'targets': targets_dict,
'epochs': n_epochs
},
execution_mode=pnl.ExecutionMode.PyTorch
)
# Print a dot for each repetition to track progress
print('.', end='')
# Print a new line every 10 repetitions
if (reps + 1) % 10 == 0:
print()
/opt/hostedtoolcache/Python/3.11.14/x64/lib/python3.11/site-packages/psyneulink/core/compositions/composition.py:10404: UserWarning: There are one or more TARGET_MECHANISMS specified in both the 'inputs' and 'targets' args of the learn() method for embedding (TARGET for context_label ,TARGET for response); This isn't technically a problem, but it is redundant so thought you should know ;^).
warnings.warn(f"There are one or more TARGET_MECHANISMS specified in both the 'inputs' and 'targets' "
---------------------------------------------------------------------------
KeyboardInterrupt Traceback (most recent call last)
Cell In[7], line 33
30 i += 1
32 # Train the network for `n_epochs`
---> 33 result = isc_ci_model.learn(
34 inputs={
35 'inputs': inputs_dict,
36 'targets': targets_dict,
37 'epochs': n_epochs
38 },
39 execution_mode=pnl.ExecutionMode.PyTorch
40 )
41 # Print a dot for each repetition to track progress
42 print('.', end='')
File /opt/hostedtoolcache/Python/3.11.14/x64/lib/python3.11/site-packages/psyneulink/core/globals/context.py:747, in handle_external_context.<locals>.decorator.<locals>.wrapper(context, *args, **kwargs)
744 pass
746 try:
--> 747 return func(*args, context=context, **kwargs)
748 except TypeError as e:
749 # context parameter may be passed as a positional arg
750 if (
751 f"{func.__name__}() got multiple values for argument"
752 not in str(e)
753 ):
File /opt/hostedtoolcache/Python/3.11.14/x64/lib/python3.11/site-packages/psyneulink/library/compositions/autodiffcomposition.py:1761, in AutodiffComposition.learn(self, synch_projection_matrices_with_torch, synch_node_variables_with_torch, synch_node_values_with_torch, synch_results_with_torch, retain_torch_trained_outputs, retain_torch_targets, retain_torch_losses, context, base_context, skip_initialization, *args, **kwargs)
1756 if execution_mode == pnlvm.ExecutionMode.PyTorch and not torch_available:
1757 raise AutodiffCompositionError(f"'{self.name}.learn()' has been called with ExecutionMode.Pytorch, "
1758 f"but Pytorch module ('torch') is not installed. "
1759 f"Please install it with `pip install torch` or `pip3 install torch`")
-> 1761 return super().learn(*args,
1762 synch_with_pnl_options=synch_with_pnl_options,
1763 retain_in_pnl_options=retain_in_pnl_options,
1764 execution_mode=execution_mode,
1765 context=context,
1766 base_context=base_context,
1767 skip_initialization=skip_initialization,
1768 **kwargs)
File /opt/hostedtoolcache/Python/3.11.14/x64/lib/python3.11/site-packages/psyneulink/core/globals/context.py:747, in handle_external_context.<locals>.decorator.<locals>.wrapper(context, *args, **kwargs)
744 pass
746 try:
--> 747 return func(*args, context=context, **kwargs)
748 except TypeError as e:
749 # context parameter may be passed as a positional arg
750 if (
751 f"{func.__name__}() got multiple values for argument"
752 not in str(e)
753 ):
File /opt/hostedtoolcache/Python/3.11.14/x64/lib/python3.11/site-packages/psyneulink/core/compositions/composition.py:12101, in Composition.learn(self, inputs, targets, num_trials, epochs, learning_rate, minibatch_size, optimizations_per_minibatch, patience, min_delta, execution_mode, randomize_minibatches, call_before_minibatch, call_after_minibatch, context, base_context, skip_initialization, *args, **kwargs)
12098 if optimizations_per_minibatch is None:
12099 optimizations_per_minibatch = self.parameters.optimizations_per_minibatch._get(context)
> 12101 result = runner.run_learning(
12102 inputs=inputs,
12103 targets=targets,
12104 num_trials=num_trials,
12105 epochs=epochs,
12106 learning_rate=learning_rate,
12107 minibatch_size=minibatch_size,
12108 optimizations_per_minibatch=optimizations_per_minibatch,
12109 patience=patience,
12110 min_delta=min_delta,
12111 randomize_minibatches=randomize_minibatches,
12112 call_before_minibatch=call_before_minibatch,
12113 call_after_minibatch=call_after_minibatch,
12114 context=context,
12115 execution_mode=execution_mode,
12116 skip_initialization=skip_initialization,
12117 *args, **kwargs)
12119 context.remove_flag(ContextFlags.LEARNING_MODE)
12120 return result
File /opt/hostedtoolcache/Python/3.11.14/x64/lib/python3.11/site-packages/psyneulink/library/compositions/compositionrunner.py:407, in CompositionRunner.run_learning(self, inputs, targets, num_trials, epochs, learning_rate, minibatch_size, optimizations_per_minibatch, patience, min_delta, randomize_minibatches, synch_with_pnl_options, retain_in_pnl_options, call_before_minibatch, call_after_minibatch, context, execution_mode, skip_initialization, **kwargs)
405 # By-pass parse learning spec if we are dealing with sequences for now
406 if not (isinstance(stim_input, list) and all(isinstance(i, dict) for i in stim_input)):
--> 407 stim_input, num_input_trials = self._composition._parse_learning_spec(inputs=stim_input,
408 targets=stim_target,
409 execution_mode=execution_mode,
410 context=context)
411 else:
412 num_trials = len(stim_input)
File /opt/hostedtoolcache/Python/3.11.14/x64/lib/python3.11/site-packages/psyneulink/library/compositions/autodiffcomposition.py:1584, in AutodiffComposition._parse_learning_spec(self, inputs, targets, execution_mode, context)
1583 def _parse_learning_spec(self, inputs, targets, execution_mode, context):
-> 1584 stim_input, num_input_trials = super()._parse_learning_spec(inputs, targets, execution_mode, context)
1586 if not callable(inputs):
1587 input_ports_for_INPUT_Nodes = self._get_input_receivers()
File /opt/hostedtoolcache/Python/3.11.14/x64/lib/python3.11/site-packages/psyneulink/core/compositions/composition.py:10411, in Composition._parse_learning_spec(self, inputs, targets, execution_mode, context)
10407 self._warned_about_targets_mechs_in_inputs_and_targets = True
10409 # 3) Resize inputs to be of the form [[[]]],
10410 # where each level corresponds to: <TRIALS <PORTS <INPUTS> > >
> 10411 inputs, num_inputs_sets = self._parse_input_dict(inputs, context)
10413 return inputs, num_inputs_sets
File /opt/hostedtoolcache/Python/3.11.14/x64/lib/python3.11/site-packages/psyneulink/core/compositions/composition.py:10573, in Composition._parse_input_dict(self, inputs, context)
10571 _inputs = self._parse_labels(_inputs)
10572 self._validate_input_dict_keys(_inputs)
> 10573 _inputs = self._instantiate_input_dict(_inputs)
10574 _inputs = self._flatten_nested_dicts(_inputs)
10575 _inputs = self._validate_input_shapes_and_expand_for_all_trials(_inputs)
File /opt/hostedtoolcache/Python/3.11.14/x64/lib/python3.11/site-packages/psyneulink/core/compositions/composition.py:10804, in Composition._instantiate_input_dict(self, inputs)
10802 else:
10803 node_name = node_spec.full_name
> 10804 error_base_msg = f"Input for '{node_name}' of '{self.name}' ({_inputs}) "
10806 if isinstance(_inputs, dict):
10807 # entry is dict for a nested Composition, which will be handled recursively
10808 pass
File /opt/hostedtoolcache/Python/3.11.14/x64/lib/python3.11/site-packages/pandas/core/series.py:1793, in Series.__repr__(self)
1791 # pylint: disable=invalid-repr-returned
1792 repr_params = fmt.get_series_repr_params()
-> 1793 return self.to_string(**repr_params)
File /opt/hostedtoolcache/Python/3.11.14/x64/lib/python3.11/site-packages/pandas/core/series.py:1880, in Series.to_string(self, buf, na_rep, float_format, header, index, length, dtype, name, max_rows, min_rows)
1827 def to_string(
1828 self,
1829 buf: FilePath | WriteBuffer[str] | None = None,
(...) 1838 min_rows: int | None = None,
1839 ) -> str | None:
1840 """
1841 Render a string representation of the Series.
1842
(...) 1878 '0 1\\n1 2\\n2 3'
1879 """
-> 1880 formatter = fmt.SeriesFormatter(
1881 self,
1882 name=name,
1883 length=length,
1884 header=header,
1885 index=index,
1886 dtype=dtype,
1887 na_rep=na_rep,
1888 float_format=float_format,
1889 min_rows=min_rows,
1890 max_rows=max_rows,
1891 )
1892 result = formatter.to_string()
1894 # catch contract violations
File /opt/hostedtoolcache/Python/3.11.14/x64/lib/python3.11/site-packages/pandas/io/formats/format.py:225, in SeriesFormatter.__init__(self, series, length, header, index, na_rep, name, float_format, dtype, max_rows, min_rows)
222 self.dtype = dtype
223 self.adj = printing.get_adjustment()
--> 225 self._chk_truncate()
File /opt/hostedtoolcache/Python/3.11.14/x64/lib/python3.11/site-packages/pandas/io/formats/format.py:247, in SeriesFormatter._chk_truncate(self)
245 else:
246 row_num = max_rows // 2
--> 247 series = concat((series.iloc[:row_num], series.iloc[-row_num:]))
248 self.tr_row_num = row_num
249 else:
File /opt/hostedtoolcache/Python/3.11.14/x64/lib/python3.11/site-packages/pandas/core/reshape/concat.py:395, in concat(objs, axis, join, ignore_index, keys, levels, names, verify_integrity, sort, copy)
380 copy = False
382 op = _Concatenator(
383 objs,
384 axis=axis,
(...) 392 sort=sort,
393 )
--> 395 return op.get_result()
File /opt/hostedtoolcache/Python/3.11.14/x64/lib/python3.11/site-packages/pandas/core/reshape/concat.py:644, in _Concatenator.get_result(self)
642 new_index = default_index(len(res))
643 else:
--> 644 new_index = self.new_axes[0]
646 mgr = type(sample._mgr).from_array(res, index=new_index)
648 result = sample._constructor_from_mgr(mgr, axes=mgr.axes)
File pandas/_libs/properties.pyx:36, in pandas._libs.properties.CachedProperty.__get__()
File /opt/hostedtoolcache/Python/3.11.14/x64/lib/python3.11/site-packages/pandas/core/reshape/concat.py:702, in _Concatenator.new_axes(self)
699 @cache_readonly
700 def new_axes(self) -> list[Index]:
701 ndim = self._get_result_dim()
--> 702 return [
703 self._get_concat_axis if i == self.bm_axis else self._get_comb_axis(i)
704 for i in range(ndim)
705 ]
File /opt/hostedtoolcache/Python/3.11.14/x64/lib/python3.11/site-packages/pandas/core/reshape/concat.py:703, in <listcomp>(.0)
699 @cache_readonly
700 def new_axes(self) -> list[Index]:
701 ndim = self._get_result_dim()
702 return [
--> 703 self._get_concat_axis if i == self.bm_axis else self._get_comb_axis(i)
704 for i in range(ndim)
705 ]
File pandas/_libs/properties.pyx:36, in pandas._libs.properties.CachedProperty.__get__()
File /opt/hostedtoolcache/Python/3.11.14/x64/lib/python3.11/site-packages/pandas/core/reshape/concat.py:760, in _Concatenator._get_concat_axis(self)
758 if self.levels is not None:
759 raise ValueError("levels supported only when keys is not None")
--> 760 concat_axis = _concat_indexes(indexes)
761 else:
762 concat_axis = _make_concat_multiindex(
763 indexes, self.keys, self.levels, self.names
764 )
File /opt/hostedtoolcache/Python/3.11.14/x64/lib/python3.11/site-packages/pandas/core/reshape/concat.py:778, in _concat_indexes(indexes)
777 def _concat_indexes(indexes) -> Index:
--> 778 return indexes[0].append(indexes[1:])
File /opt/hostedtoolcache/Python/3.11.14/x64/lib/python3.11/site-packages/pandas/core/indexes/base.py:5501, in Index.append(self, other)
5498 names = {obj.name for obj in to_concat}
5499 name = None if len(names) > 1 else self.name
-> 5501 return self._concat(to_concat, name)
File /opt/hostedtoolcache/Python/3.11.14/x64/lib/python3.11/site-packages/pandas/core/indexes/base.py:5511, in Index._concat(self, to_concat, name)
5507 to_concat_vals = [x._values for x in to_concat]
5509 result = concat_compat(to_concat_vals)
-> 5511 return Index._with_infer(result, name=name)
File /opt/hostedtoolcache/Python/3.11.14/x64/lib/python3.11/site-packages/pandas/core/indexes/base.py:675, in Index._with_infer(cls, *args, **kwargs)
671 result._references.add_index_reference(result)
673 return result
--> 675 @classmethod
676 def _with_infer(cls, *args, **kwargs):
677 """
678 Constructor that uses the 1.0.x behavior inferring numeric dtypes
679 for ndarray[object] inputs.
680 """
681 result = cls(*args, **kwargs)
KeyboardInterrupt:
Testing#
embedding_falcon = embeddings_df.loc['falcon']
embedding_owl = embeddings_df.loc['owl']
embedding_cat = embeddings_df.loc['cat']
embedding_penguin = embeddings_df.loc['penguin']
res_cat = isc_ci_model.run(
{
query: embedding_cat,
support_1: embedding_falcon,
support_2: embedding_owl,
})
response.value
array([[0.4971299 , 0.50272648]])
res_penguin = isc_ci_model.run(
{
query: embedding_penguin,
support_1: embedding_falcon,
support_2: embedding_owl,
})
response.value
array([[0.47386775, 0.52613219]])
Since the training was not sufficient, the model will not be able to predict the correct response. However, can you explain what the expected outcome for the above support and query items would be?
✅ Solution
The expected outcome for the above support and query items would be:
Higher split between yes and no for the query cat than for the query penguin.
Even with a larger training set, the model is unlikely to “see” each combination of support and query items for each possible context. How can it still learn contexts for unseen combinations?
💡 Hint
Why are we using context independent layers in the first place? What would happen if we just use one-hot encodings and skip these independent layer?
✅ Solution
The embeddings make it so that the model generalizes to unseen combinations since they might be “similar” to seen combinations. For example, birds share features in embedding space. So even if the model hasn’t seen specific bird combinations it still learns the context that is elicited by these combinations.
If we just use one-hot encodings and skip the context independent layer, the model would not be able to generalize to unseen combinations since they don’t share any activations (everything is orthogonal). This would mean that with enough training, the model would maybe still learn to make correct predictions but never on unseen combinations.